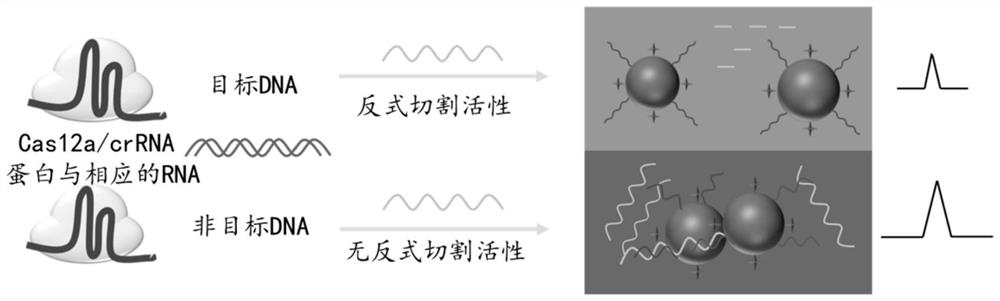
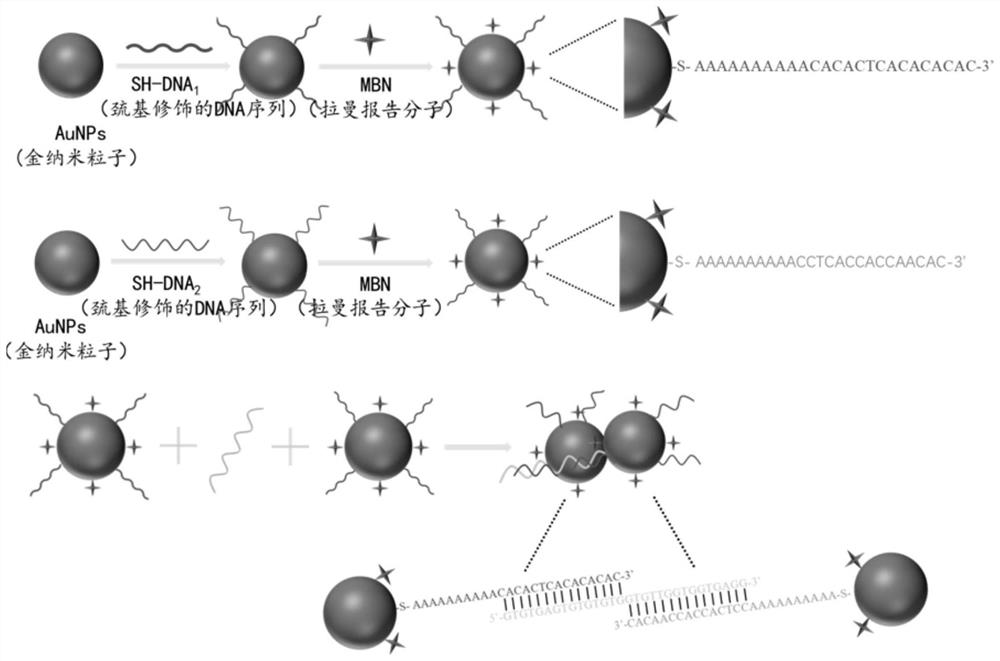
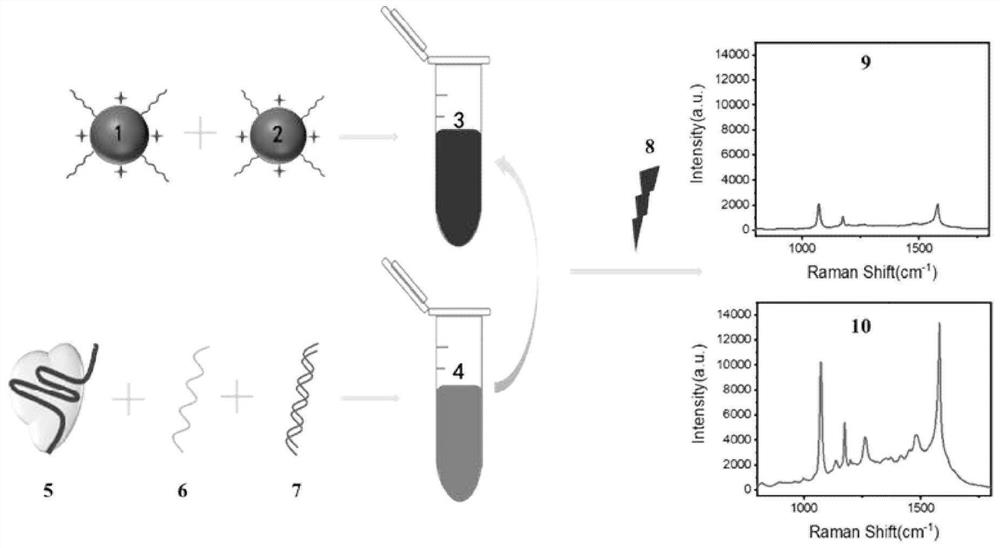
Surface enhanced Raman spectrum detection method for gene based on CRISPR/Cas12a protein
A surface-enhanced Raman and spectral detection technology, which is applied in the field of spectral detection of virus genes, can solve the problems of weakening and SERS signal enhancement, and achieve the effects of high success rate, high sensitivity detection, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 CRISPR / Cas12a combined with SERS technology for detection of HPV16 nucleic acid
[0053] In this embodiment, the metal nanoparticle probe is a gold nanoparticle probe.
[0054] In this embodiment, the Raman signaling molecule is MBN.
[0055] In this example, the DNA 1 The sequence is SH-AAAAAAAAAAACACACTCACACACAC.
[0056] In this example, the DNA 2 The sequence is SH-AAAAAAAAAACCTCACCACCAACAC.
[0057] In this embodiment, the sequence of the substrate ssDNA is GTGTGAGTGTGTGTGGTGTTGGTGGTGAGG.
[0058] 在该实施例中,所述靶标核酸为HPV 16 DNA具体序列为CTATGGATTACAAACAAACACAATTGTGTTTAATTGGTTGCAAACCACCTATAGGGGAACACTGGGGCAAAGGATCCCCATGTACCAATGTTGCAGTAAATCCAGGTGATTGTCCACCATTAGAGTTAATAAACACAGTTATTCAGGATGGTGATATGGTTGATACTGGCTTTGGTGCTATGGACTTTACTACATTACAGGCTAACAAAAGTGAAGTTCCACTGGATATTTGTACATCTA。
[0059] In this embodiment, the crRNA is the corresponding crRNA of HPV 16 16 , the specific sequence is UAAUUUCUACUAAGUGUAGAUCUACAUUACAGGCUAACAAA.
[0060] (1) Preparation of gold nanopa...
Embodiment 2
[0083] Example 2 CRISPR / Cas12a combined with SERS technology for detection of HPV18 nucleic acid
[0084] In this embodiment, the metal nanoparticle probe is a gold nanoparticle probe.
[0085] In this embodiment, the Raman signaling molecule is MBN.
[0086] In this example, the DNA 1 The sequence is SH-AAAAAAAAAAACACACTCACACACAC.
[0087] In this example, the DNA 2 The sequence is SH-AAAAAAAAAACCTCACCACCAACAC.
[0088] In this embodiment, the sequence of the substrate ssDNA is GTGTGAGTGTGTGTGGTGTTGGTGGTGAGG.
[0089] 在该实施例中,所述靶标核酸为HPV 18 DNA片段,具体序列为GGGAACACTGGGCTAAAGGCACTGCTTGTAAATCGCGTCCTTTATCACAGGGCGATTGCCCCCCTTTAGAACTTAAAAACACAGTTTTGGAAGATGGTGATATGGTAGATACTGGATATGGTGCCATGGACTTTAGTACATTGCAAGATACTAAATGTGAGGTACCATTGGATATTTGTCAGTCTATTTGTAAATATCCTGATTATTTACAAATGTCTGCAGATCCTTATGGGGATTCCA。
[0090] In this embodiment, the crRNA is HPV 18 crRNA 18 , the specific sequence is UAAUUUCUACUAAGUGUAGAUGUACAUUGCAAGAUACUAAA.
[0091] Steps (1) and (2) to prepare gold nanoparticles ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com