Construction of threonine production strain and method for producing threonine
A construction method and threonine technology, which is applied in the construction of threonine production strains with high conversion rate and the field of threonine production, can solve the problems of multiple by-products, slow growth of strains, difficulty in obtaining high-yield strains, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
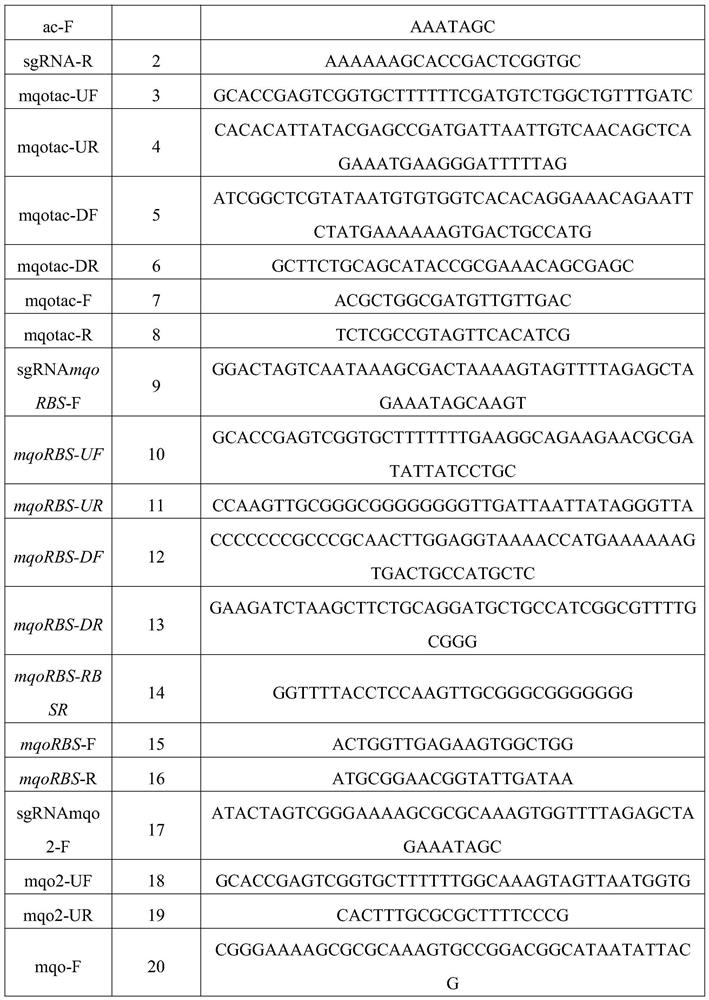
[0036] Example 1: Preparation of strain MHZ-0214-3 (promoter strengthening) that strengthens the mqo gene
[0037] (1) pTarget-tac-mqo plasmid construction
[0038] Step1: Using the pTargetT plasmid as a template (from the document Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System, JiangY, Chen B, et al. Appl. Environ Microbiol, 2015), select the sgRNAmqotac-F / sgRNA-R primer pair, The sgRNA fragment with N20 was amplified①; Step2: Using the W3110 genome as a template, the mqotac-UF / mqotac-UR primer pair was selected to amplify the upstream homology arm containing the tac promoter②; Step3: Using the W3110 genome as a template As a template, select the mqotac-DF / mqotac-DR primer pair to amplify the downstream homology arm ③ containing the tac promoter; Step4: use ①, ②, ③ as templates, select the sgRNAmqotac-F / mqotac-DR primer pair, and amplify Add the sgRNA-up-Ptac-down fragment, also known as Donor DNA; Step5: Carry out SpeI and PstI double enzyme dig...
Embodiment 2
[0044] Embodiment 2: the bacterial strain MHZ-0214-4 (RBS-mqo) of the mqo gene that prepares RBS strengthening
[0045] (1) pTarget-RBS3-mqo plasmid construction
[0046] Step1: Using the pTargetT plasmid as a template (from the document Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System, JiangY, Chen B, et al. Appl. Environ Microbiol, 2015), select the sgRNAmqoRBS-F / sgRNA-R primer pair, The sgRNA fragment with N20 was amplified ①; Step 2: Using the W3110 genome as a template, the mqoRBS-UF / mqoRBS-UR primer pair was selected to amplify the upstream homology arm containing RBS ②; Step 3: Using the fragment ② as a template, Use the mqoRBS-UF / mqoRBS-RBSR primer pair to amplify the upstream homology arm containing RBS③; Step4: Using the W3110 genome as a template, use the mqoRBS-DF / mqoRBS-DR primer pair to amplify the downstream homology arm containing RBS Source arm ④; Step5: ①, ③, ④ as the template, select the sgRNAmqoRBS-F / mqoRBS-DR primer pair, amplif...
Embodiment 3
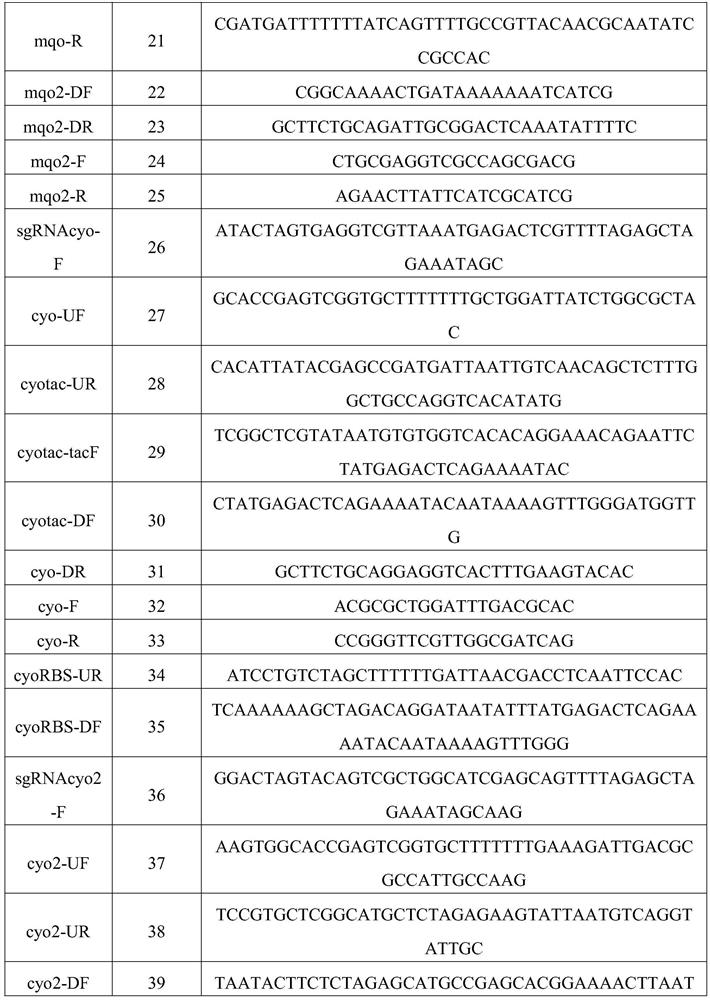
[0053] Embodiment 3: prepare the bacterial strain MHZ-0214-5 (multiple copy) of strengthening mqo gene
[0054] (1) pTarget-zwf::mqo plasmid construction
[0055] Step1: Using the pTargetT plasmid as a template (from the literature Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System, JiangY, Chen B, et al. Appl. Environ Microbiol, 2015), select the sgRNAmqo2-F / sgRNA-R primer pair to amplify Add the sgRNA fragment with N20①; Step2: Use the W3110 genome as a template, select the mqo2-UF / mqo2-UR primer pair, and amplify the upstream homology arm②; Step3: Use the W3110 genome as a template, select mqo-F / mqo-R primer pair to amplify the mqo fragment ③; Step4: using the W3110 genome as a template, select the mqo2-DF / mqo2-DR primer pair to amplify the downstream homology arm containing RBS ④; Step5: use ①, ②, ③, ④ are used as templates, and sgRNAmqo2-F / mqo2-DR primer pair is selected to amplify the sgRNA-up-mqo-down fragment, also called DonorDNA; Step6: Spe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com