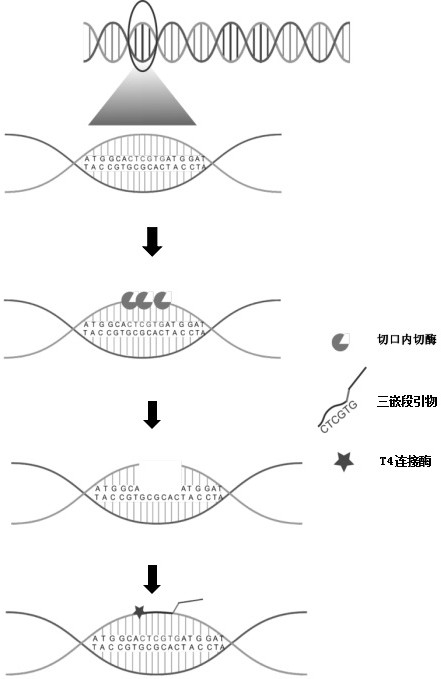
Non-diagnostic gene localization method
A gene positioning and purpose technology, applied in the field of detection analysis and biological application, can solve the problems of time-consuming, low accuracy, high cost, etc., and achieve the effect of shortening the detection time and good biocompatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Positioning method using only Nb.BssSI digestion and triangular origami probe labeling
[0071] The PCR amplification process of the target sequence in step S1, its concrete operation is:
[0072] PCR reaction of linear DNA: the sequences of the upstream and downstream primers in this process are designed by the software Primer-BLAST. The 50 μL reaction system contains: 10 μL 5×LongAmp Taq reaction solution, 2 μL dNTP mixture (2.5 mM), 1 μL LongAmp Taq DNA polymerase, 4 μL upstream primer (10 μM), 4 μL downstream primer (10 μM), 1.5 μL LambdaDNA, 27.5 μL ultrapure water. The components were mixed quickly on ice and transferred to a PCR machine. Set the PCR reaction program as follows: ① 95 °C, 2 min; ② 98 °C, 10 s; 42 °C, 50 s; 72 °C, 7 min (set 30 cycles); ③ 72 °C, 10 min; slowly cool down to 4 °C for storage. The nucleotide sequence of the Lambda DNA is provided in GenBank Accession No. J02459.1; the full length of the Lambda DNA is 48502 bp, the upstrea...
Embodiment 2
[0081] Example 2: Positioning method using only Nb.BbvCI enzyme digestion and cross-shaped origami probe labeling
[0082] Embodiment 2 except that following steps are different, all the other operations are identical with embodiment 1:
[0083] The enzyme digestion system used in step S2 is: Nb.BbvCI digestion into 30 uL system contains: 1 uL Nb.BbvCI, 3 uLRutSmart™ buffer, 26 uL of the DNA obtained in step S1;
[0084] In step S3, use the triblock primer and T4 DNA Ligase to fill the "gap", the nucleotide sequence of the triblock primer is shown in sequence NO: 8, sequence NO: 8: GCTGAGGTTTTTAAAAAAAAAAA; according to Example 1 Atomic force microscopy characterization in , the results are as follows Figure 4c and 4d as shown, Figure 4c It is a schematic diagram and an atomic force microscope characterization map of gene localization using triangular origami probes after digestion with Nb.BbvC on the short double-stranded template strand after linear double-stranded Lambd...
Embodiment 3
[0086] Example 3: A gene localization method using two nicking endonucleases and corresponding origami probe markers at the same time, the specific operations are as follows:
[0087](1) Restriction endonuclease digestion reaction: This process includes two restriction endonuclease digestion reactions, and the digestion is 30uL. The system contains: 1 uL Nb.BbvCI, 1 uL Nb.BssSI, 3 uL NEBuffer™ r3.1, 25 uL The DNA obtained in step 1 was incubated at 37°C for 6 h, then inactivated at 80°C, and then purified with TaKaRa MiniBEST DNA Fragment Purification Kit Ver.4.0 kit. Since Nb.BbvCI and Nb.BssSI are in NEBuffer™ The activity in r3.1 is 100%, so the above reaction buffer is selected as NEBuffer™r3.1.
[0088] (2) PCR reaction to fill the "gap" DNA: 28 uL reaction system contains: 3 uL T4 DNA LigaseReaction Buffer, 2 uL 1X TA-Mg 2+ Buffer, 23 uL "gapped" DNA. The components were mixed quickly on ice and transferred to a PCR machine. Set the PCR reaction program as follows: ① ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com