Method for detecting JC polyoma virus through real-time fluorescent quantitative PCR (Polymerase Chain Reaction)
A real-time fluorescence quantitative, polyoma virus technology, applied in the field of biomedicine, can solve the problem of low coverage of JC virus variant strains, and achieve the effects of reducing labor and material costs, high sensitivity, and ensuring safety.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
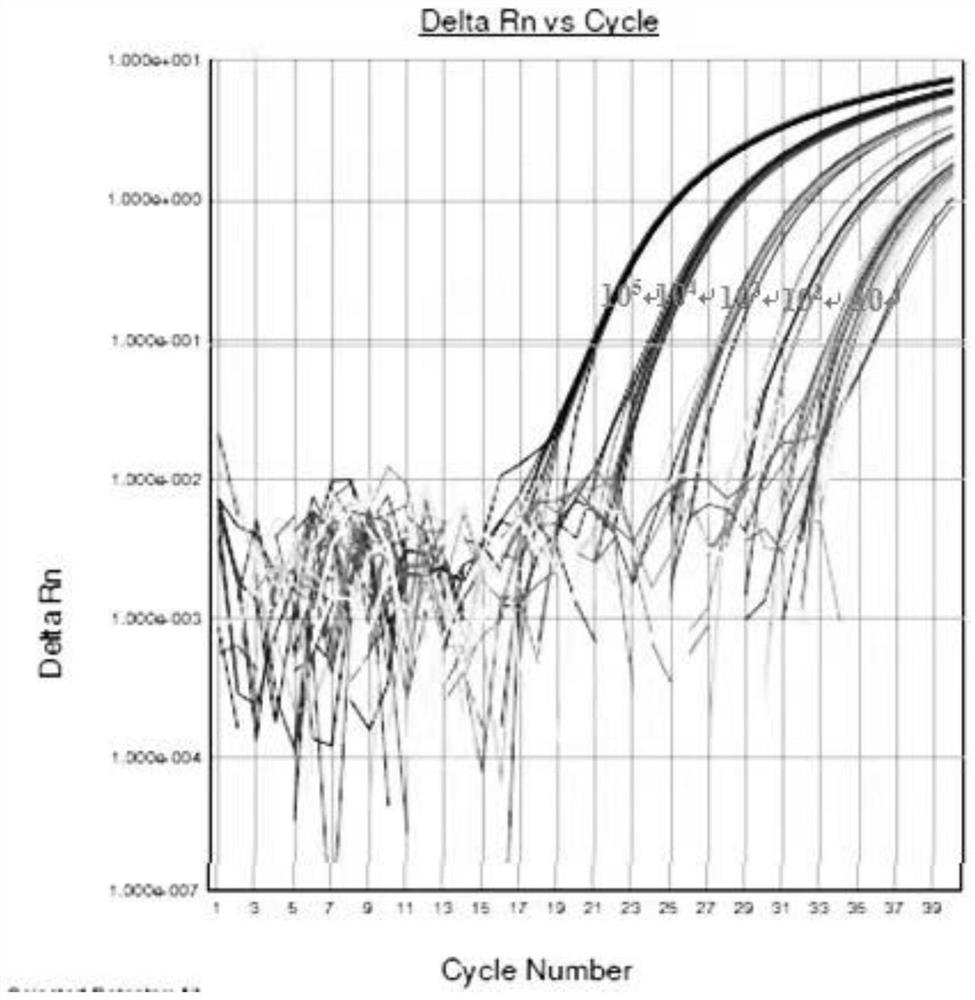
Image
Examples
Embodiment 1
[0032] 1. Extraction of the cells to extract the total DNA to be tested, and adjusted the concentration of the DNA template to 0.1 μg / μL.
[0033] 2. For all JC virus separation plant genomic DNA sequences in the NCBI database, a positive primer, reverse primer, and probe of real-time fluorescence quantitative PCR are designed.
[0034] The nucleotide sequence of forward primers is: acagagcacaaggcgtac (shown in SEQ ID NO: 1);
[0035] The nucleotide sequence of reverse primers is: Tcctcctgttagtgtcccaaa (as shown in SEQ ID NO: 2);
[0036] The nucleotide sequence of the probe is: atcctgttgaatgtggggttcctgatcc (shown in SEQ ID NO: 3).
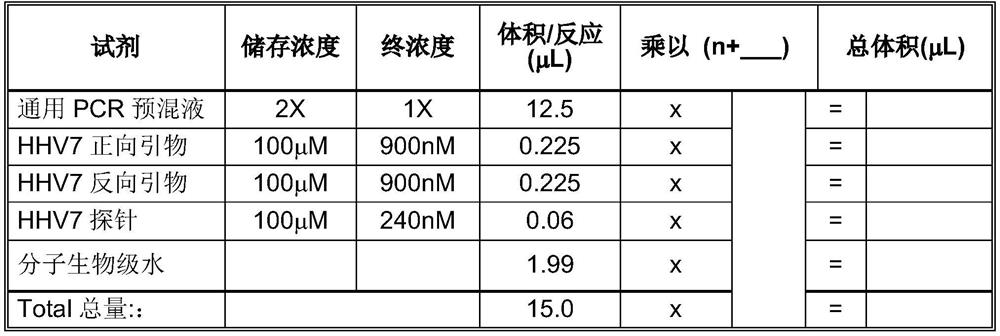
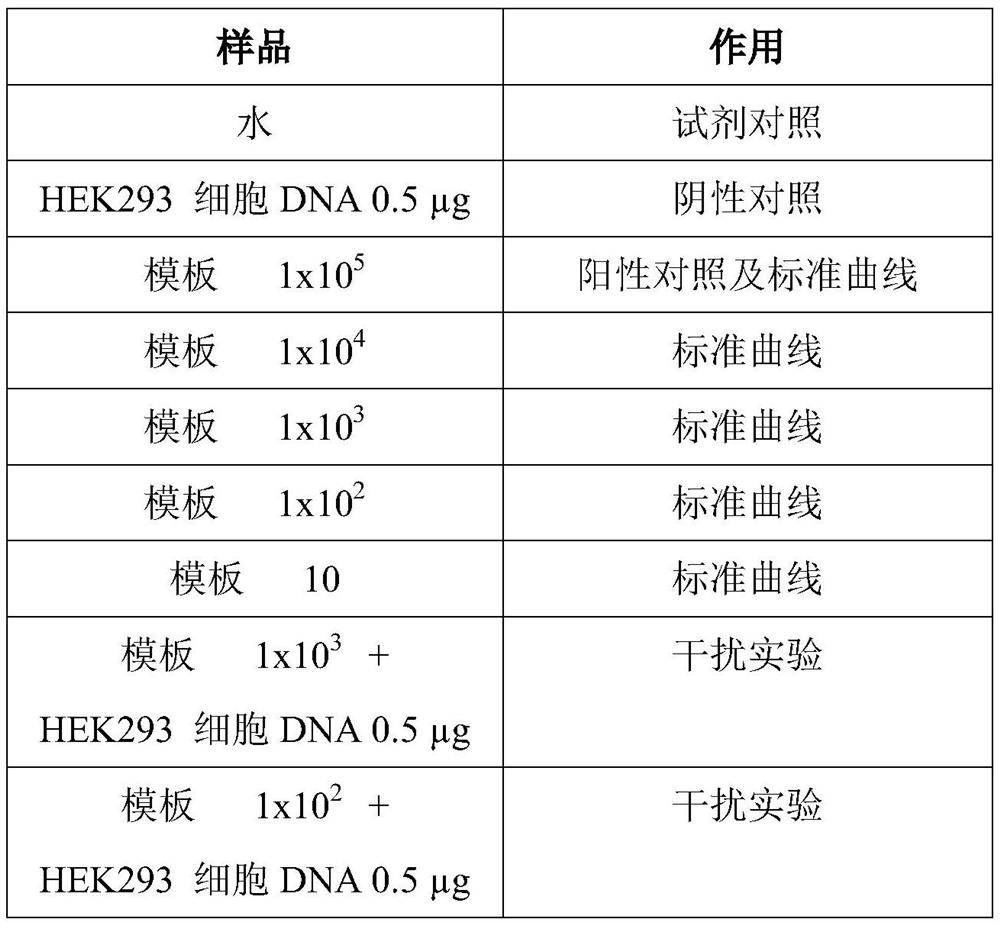
[0037] 3. Calculate the reactive number N of the real-time fluorescent quantitative PCR, and prepare the reaction system, the reaction system includes a premix, a forward primer, a reverse primer, a probe, and a template, and the premix is TAQMAN UNIVERSAL MASTER MIX, and the template is a template target sequence. The preparation of the plasmid a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com