M-MLV reverse transcriptase and preparation method thereof
A technology of reverse transcriptase and reverse transcription, which is applied in the field of biological enzymes, can solve the problems of poor thermal stability, weak ability to recognize RNA templates, and low stability, and achieve high yield, high purity, and good thermal stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
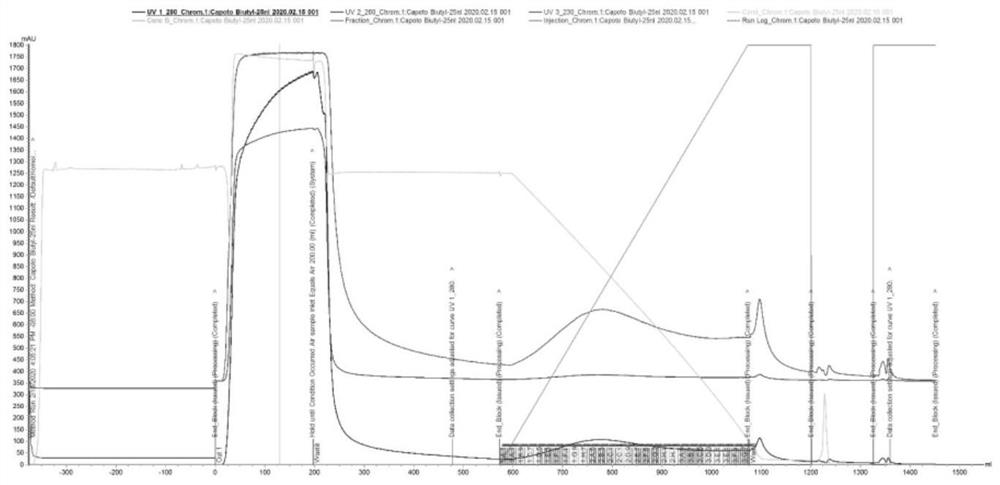
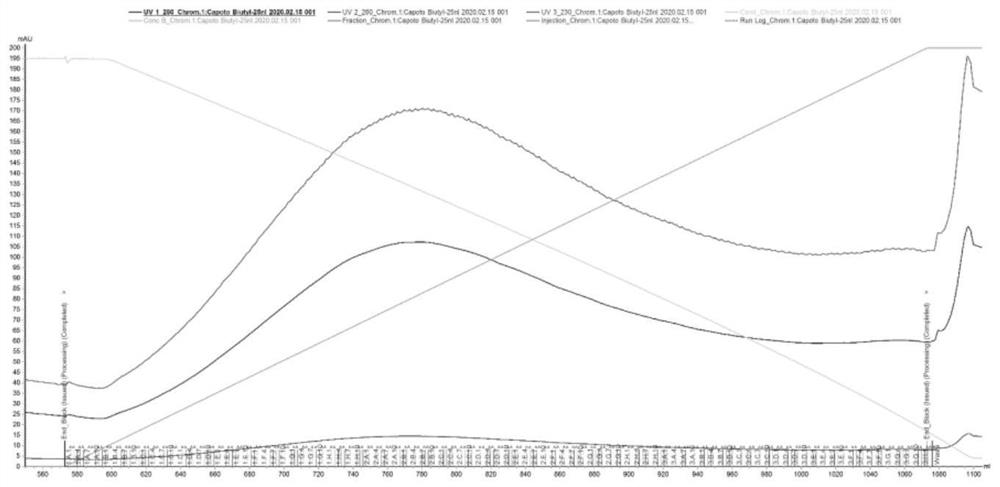
Image
Examples
Embodiment 1
[0064] construct vector
[0065] 1) Synthesize the gene sequence according to SEQ ID NO:1 and insert the whole gene fragment into the vector pET-28a (XhoI and XbaI) vector. The pET-28a expression vector was used in this example, but it is not limited to this vector.
[0066] 2) According to the positions of the mutation points D524G, E562Q, D538N, and G638H, modify the DNA sequence so that the corresponding amino acid changes, which can be modified to one or more mutations of D524G, E562Q, D538N, and G638H.
[0067] 3) Those skilled in the art fully understand that since the same amino acid may be determined by multiple different codons, the construction of a synthetic gene sequence corresponding to the mutation of the above amino acid site is not limited to one, but may be multiple cores Nucleotides are synonymously mutated to obtain a nucleotide sequence that can also encode the amino acid sequence of the mutant described in the present invention, or a nucleotide sequence c...
Embodiment 2
[0069] induced expression
[0070] Select the recombinant M-MLV plasmid that has been modified at all four sites D524G, E562Q, D538N, and G638H in Example 1.
[0071] Transformation experiment: Take a C3029 / C2529 competent cell and place it on ice for 20 minutes, and wait until the competent cell is thawed into a liquid state; take 1 μL of 10 ng / μL M-MLV plasmid and add it to the thawed competent cell (50-100 μL), Gently flick twice to mix well, rest on ice for 25 minutes; heat shock in 42°C water bath for 90 seconds, then quickly ice-bath for 5 minutes; add 1000 μL of fresh anti-antibiotic-free LB liquid medium, place on a shaker at 200 rpm and incubate at 37°C for 1 hour;
[0072] Take 200μL and spread evenly on Kan + Incubate the resistant LB plate (0.1%) in a constant temperature incubator at 37°C for 16 hours.
[0073] Induced expression: Pick the monoclonal colonies with normal growth in the plate to 150mL Kan + Resistant LB liquid medium (100μg / mL Kan + , stock solu...
Embodiment 3
[0075] Bacteria treatment
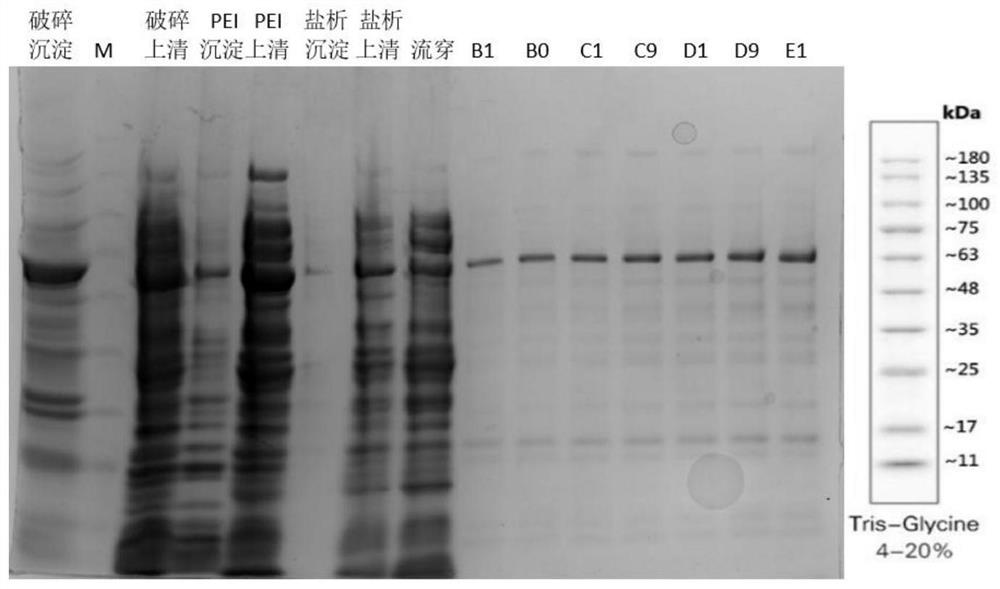
[0076] High-pressure crushing: use an electronic balance to weigh 20g of wet bacteria, add 30mL M-MLV Ni BindingBuffer (20mM Tris-HCl, 500mM NaCl, pH=8.0@25℃), and use a vortex mixer to quickly vortex until No obvious bacterial clumps were observed with the naked eye. Continue to add M-MLV Ni Binding Buffer and vortex until no obvious bacterial clumps were observed with the naked eye. Finally, use M-MLV Ni Binding Buffer to dilute to 200mL and place in an ice bath for later use. The cold trap of the high-pressure cell disruptor was pre-cooled to 4°C in advance, drained 75% ethanol in the pipeline, added 500mL process water to clean the pipeline, then added 200mL M-MLV Ni Binding Buffer to rinse the pipeline, drained it, and poured it into a heavy Suspension liquid, 1200bar, flow rate adjusted to 50, broken 6 times, cell broken liquid placed in ice bath for temporary storage. After pressure relief, add 500mL of process water to flush the pipeline, a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com