Escherichia coli for synthesizing monophosphate lipoid A containing only three fatty acid chains
A technology for Escherichia coli and lipids, which is applied in the fields of genetic engineering and bioengineering, and can solve the problems that lipid A has a large molecular weight and cannot be used to identify lipid A acyltransferases and multi-structural components.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Construction of embodiment 1 knockout plasmid
[0038] Using the CRISPR / Cas9 knockout system to knock out three genes related to lipid A secondary acyl chain transferase in E. coli requires the construction of three knockout plasmids: pTargetF-lpxL, pTargetF-lpxP and pTargetF-lpxM, these plasmids The construction process is as follows:
[0039] (1) Select 20nt N complementary to the target sequence of the target gene 20 sequence, specific N 20 The sequences are the underlined sequences of the primers pTargetF-lpxL, pTargetF-lpxP and pTargetF-lpxM respectively, and these sequences are modified to the 5' ends of the forward and reverse primers of the plasmid pTargetF to obtain the forward primers pTargetF-lpxL-F, pTargetF-lpxP- F, pTargetF-lpxM-F and reverse primers pTargetF-lpxL-R, pTargetF-lpxP-R, pTargetF-lpxM-R.
[0040] Using the plasmid pTargetF as a template, the forward and reverse primers of pTargetF-lpxL, pTargetF-lpxP and pTargetF-lpxM were used for PCR ampl...
Embodiment 2
[0044] Example 2 Construction of Escherichia coli lipid A structure-reduced strain HWJ003
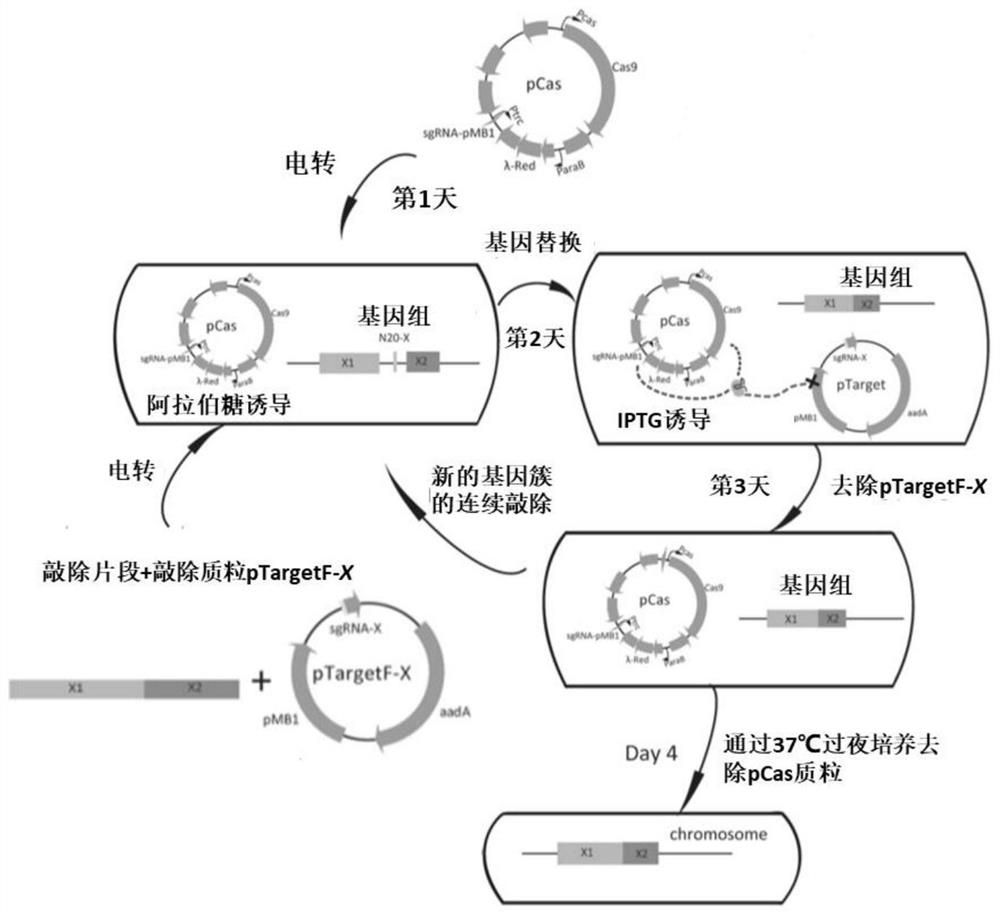
[0045] The CRISPR / Cas9 knockout system was used to knock out the lipid A secondary acyl chain transferase-related genes of Escherichia coli HW003. The specific knockout process is as follows ( figure 1 ):
[0046] (1) Preparation of Escherichia coli electroporation knockout competent cells HW003 / pCas
[0047] The plasmid pCas is transformed into Escherichia coli HW003 to obtain recombinant Escherichia coli HW003 / pCas containing the pCas plasmid, and the Escherichia coli HW003 / pCas is activated on the LB solid plate adding 30mg / L Kanamycin (Kan), and inoculated into LB (Kan+) test tube was cultivated overnight to obtain seed liquid; the seed liquid was transferred to 25mL LB (Kan+) medium at 1% (v / v), and cultivated to OD at 30°C and 200rpm 600 =0.2, add 500 μL L-arabinose solution to induce, continue to culture to OD 600 =0.5, ice bath for 30min; 4°C, centrifuge at 4000rpm for 10min...
Embodiment 3
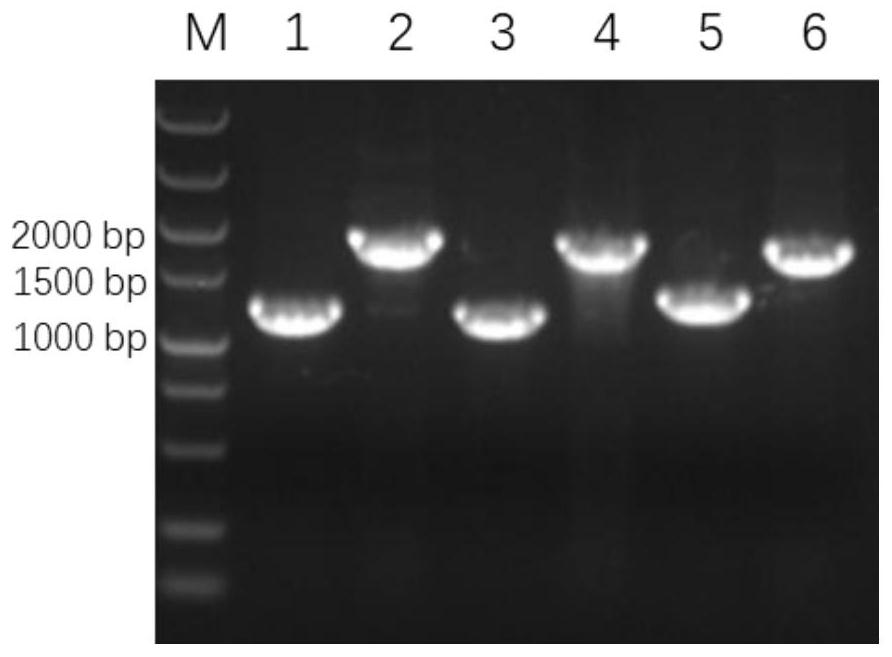
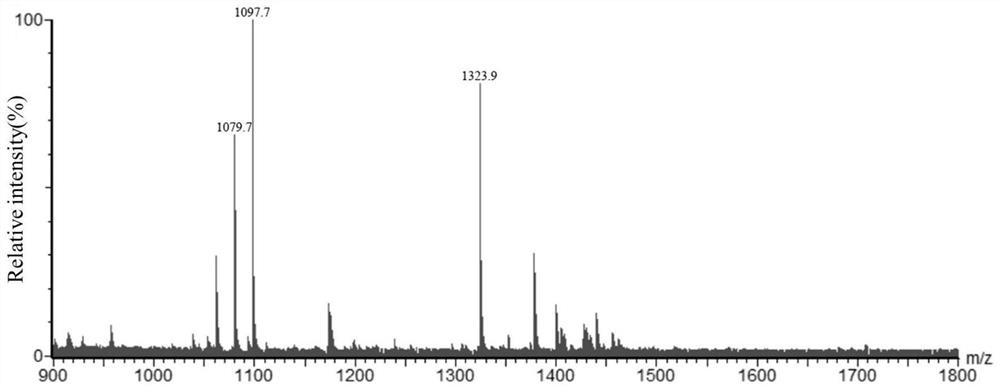
[0057] Example 3 Extraction and Verification of HWJ003 Lipid A
[0058] (1) Lipid A extraction and purification method: the extraction of lipid A adopts the mixed phase extraction method of chloroform / methanol / water solution. The strain HWJ003 was inoculated in LB liquid medium and cultured overnight at 37°C to obtain the bacterial liquid, and the initial OD of the bacterial liquid was 600 =0.02 Transfer to 1L LB liquid medium, cultivate to the late logarithmic period of the bacteria at 37°C, collect the bacteria by centrifugation at 4000rpm for 20min, and use Bligh-Dyer one-phase system chloroform / H 2 O / methanol (1:0.8:2v / v / v) 76mL mixture to suspend the cell pellet, magnetically stir for 1h, centrifuge at 2000rpm for 30min to collect the cell debris, add 27mL sodium acetate (pH 4.5) to the precipitate, and ultrasonically homogenize for 10min , and heated in a boiling water bath for 30min. The suspension was mixed with 60mL of methanol / chloroform (1:1v / v) to form a Bligh-Dy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com