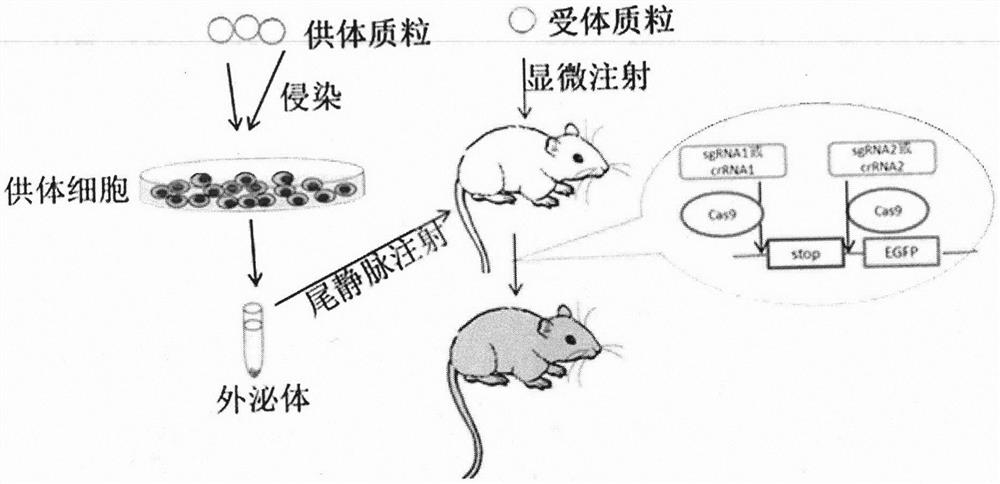
Construction and application of transgenic mouse model for exosome tracing
A technology of transgenic mice and exosomes, applied in the construction of transgenic mouse models, can solve the problems of silencing, reading frame changes, loss of function of target genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
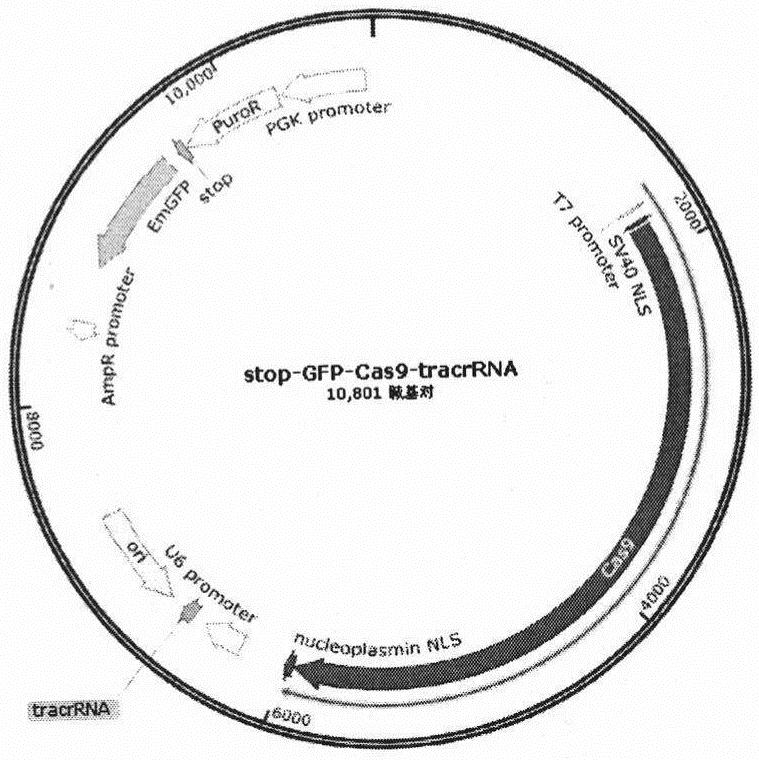
[0019] Example 1 Construction of a plasmid expressing stop-EGFP-Cas9-tracrRNA element (exosome receptor plasmid)
[0020] Construct a plasmid expressing the stop-EGFP-Cas9-tracrRNA element. The plasmid contains elements such as stop-EGFP-Cas9-tracrRNA for constructing exosome receptors; the plasmid vector contains the Cas9 protein sequence, and we need to insert it into the plasmid vector The stop-EGFP sequence and the tracrRNA sequence are sufficient. The specific operation method is as follows
[0021] Double-enzyme digestion of the plasmid vector, run 1% agarose gel and gel to recover larger gene fragments; complete gene synthesis of stop-EGFP DNA sequence and tracrRNA DNA sequence; double-enzyme cut double-stranded stop-EGFP sequence, pass through the column to recover the enzyme Cut the product, then connect the double-enzyme-cut vector and fragment, transform it into DH5α, select bacteria and sequence, and screen out the successfully constructed clone; cut the correctly ...
Embodiment 2
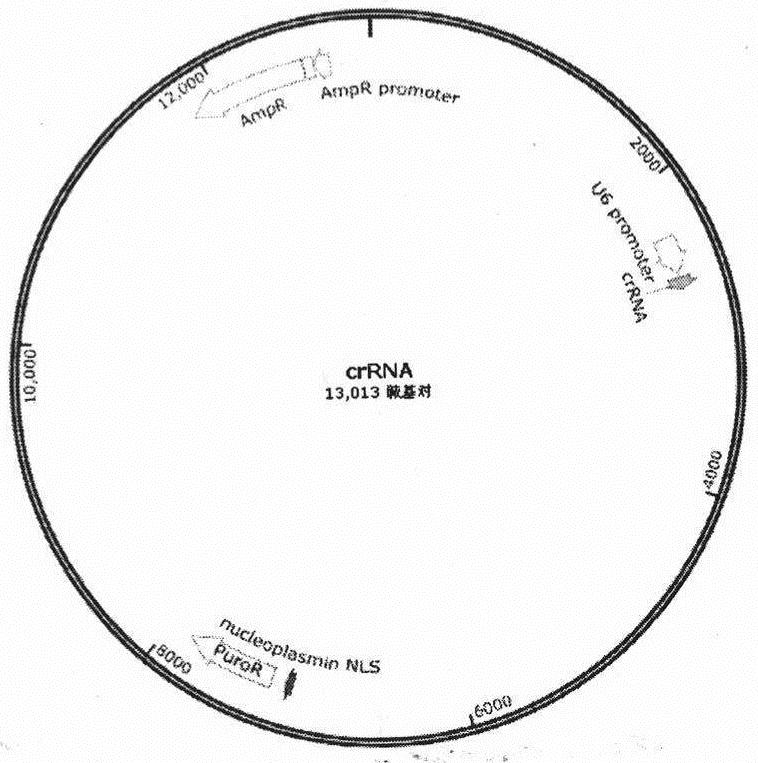
[0022] Example 2 Construction of a plasmid expressing crRNA or sgRNA (exosome donor plasmid)
[0023] Construct a plasmid expressing crRNA or sgRNA, the plasmid contains crRNA-1 / crRNA-2 or sgRNA-1 / sgRNA-2; used to construct an exosome donor; insert the crRNA / sgRNA sequence behind the U6 promoter of the plasmid vector Yes, the specific operations are as follows:
[0024] Double-enzyme digestion of the plasmid vector, run 1% agarose gel and gel to recover larger gene fragments, design crRNA-1 and crRNA-2 sequences targeting stop sequences and anneal them into oligo double-stranded in vitro. The crRNA-2 gene fragment was enzymatically linked with the double-enzyme-digested plasmid vector, and after transformation and sequencing, the successfully constructed clone was screened.
Embodiment 3
[0025] Example 3 Construction of Donor Cells and Verification of Exosomes
[0026] The construction of the donor cells is divided into two parts. The first part uses the tool cell HEK 293T cells to prepare the lentivirus expressing the donor element and obtains the virus suspension. The second part uses the obtained virus suspension to infect human non-small cell lung cancer. A549 cells, after screening, pick out the stable transfection monoclonal cell line, expand the culture to collect exosomes and detect; the specific operations are as follows
[0027] 1. Mix Opti-MEM, Lipo2000 / PEI, the target plasmid and the helper plasmid according to a certain ratio and let it stand for 20 minutes, then slowly drop it into a HEK 293T cell culture dish with a growth density of 70% to 80%, change it after 6 hours After 48 hours, the cell culture medium (containing lentivirus) was collected in a biological safety cabinet, and filtered with a 0.45 μm filter to obtain the virus supernatant, w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com