Corn naringenin methyltransferase gene ZmNOMT and application thereof in plant broad-spectrum disease resistance
A technology of prime methyl and transferase, applied in the direction of transferase, application, plant products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
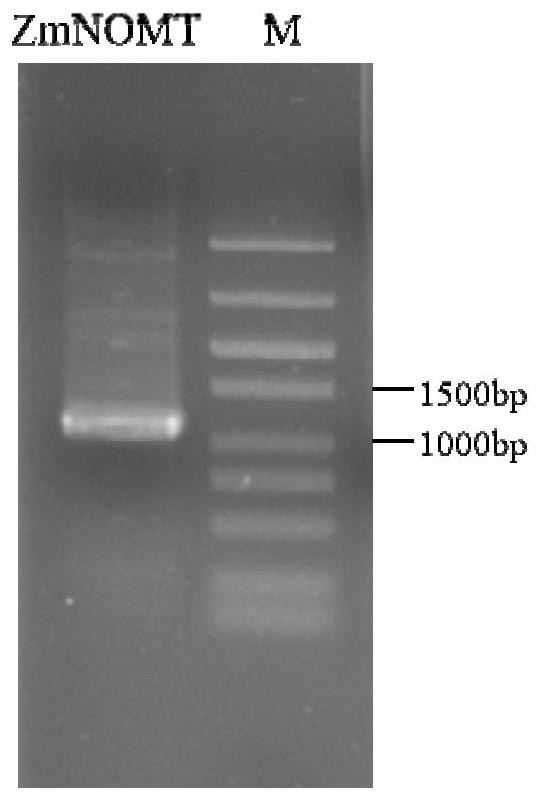
[0049] Example 1. Cloning of ZmNOMT
[0050] 1.1 Extraction of total RNA from maize inbred line B73
[0051] (1) Put the corn B73 leaf material into a high-temperature sterilized mortar, pour it into liquid nitrogen and grind it into powder; continuously add liquid nitrogen during the grinding process to prevent melting;
[0052] (2) Transfer the ground leaf powder into a 1.5ml centrifuge tube, quickly add 1ml RNAiso Plus extract, vortex, and let stand for 10min at room temperature;
[0053] (3) Add 200 μl of chloroform, gently shake and mix until the liquid is stratified, and leave it at room temperature for 5 minutes;
[0054] (4) 4°C, 12,000rpm, centrifuge for 15min, transfer 500μl of supernatant to a new 1.5ml centrifuge tube, add 500μl of isopropanol, invert up and down to mix, and place at room temperature for 10mim;
[0055] (5) 4°C, 12,000rpm, centrifuge for 15min, discard the supernatant, wash the precipitate with 1ml 75% ethanol, flick the bottom of the tube to mak...
Embodiment 2
[0087] Example 2. Construction of ZmNOMT overexpression vector and verification of disease resistance function of transgenic lines
[0088] 2.1 Construction of maize ZmNOMT overexpression vector
[0089] (1) Double enzyme digestion of maize overexpression vector pCAMBIA3301 with BamH1 and Xma1;
[0090] (2) BamH1 and Xma1 restriction sites were added at both ends of the gene ZmNOMT;
[0091] (3) The target band is recovered by gel after double enzyme digestion of the gene, and connected to the pCAMBIA3301 carrier through T4 DNA ligase;
[0092] (4) Transform E. coli competent DH5α, identify the positive clones by colony PCR, and sequence the plasmid in advance. The plasmid vector after correct sequencing is named pCAMBIA3301::ZmNOMT, and its map is shown in Figure 4 .
[0093] 2.2 Plasmid transformation Agrobacterium
[0094] (1) Add 1 μg of plasmid vector to 50 μl of Agrobacterium competent EHA105, mix well, and take an ice bath for 30 minutes;
[0095] (2) Quick-freeze...
Embodiment 3
[0151] Example 3. Construction of ZmNOMT Gene Editing Vector and Verification of Disease Resistance of Edited Lines
[0152] 3.1 Construction of maize ZmNOMT gene editing vector
[0153] The CRISPR / CAS9 base vector was single digested with HindIII. Based on the principle of CRISPR / CAS9 technology and ZmNOMT genome sequence, two target sites, gRNA1 and gRNA2, were designed. The nucleotide sequence of gRNA1 is shown in SEQ ID No.3, and the nucleotide sequence of gRNA2 is shown in SEQ ID No.4. Show. The U-S (U6-gRNA-sgRNA cassette) fragment was used as the template to design specific amplification primers with vector nick homology arms and overlapping PCR overlapping parts (20bp gRNA part), and U-S was used as the template to amplify the target fragment U6-2 promoter And sgRNA scanfold fragment, finally obtained CPB::U6-2::gRNA::sgRNA::CPB overlapping fragment by overlapping PCR, after the overlapping PCR product was sequenced correctly, T4 DNA ligase (TaKaRa) was used to conne...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com