High affinity nanoantibodies of SARS-CoV-2 alpha, gamma, delta and o mutants camel source
A technology for sars-cov-2 and mutant strains, which is applied in the field of camel-derived high-affinity nanobodies for SARS-CoV-2 α, γ, δ, and ο mutant strains, and can solve the problem of difficulty in obtaining plasma and the small amount of plasma from patients who have recovered from serum therapy problem, to achieve the effect of high stability, high neutrality and ability, high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1. Construction of SARS-CoV-2 Nanobody Library
[0086] Take 200ug of SARS-CoV-2 virus Wild Type original strain S protein and RBD protein (Beijing Yiqiao Shenzhou Biological Co., Ltd.) mixed with an equal volume of complete Freund's adjuvant, fully emulsified and injected into camels, and then boosted immunization every two weeks One time, the mixture of incomplete Freund's adjuvant and immunogen was used in booster immunization, and multiple subcutaneous immunizations on the back of the neck were used for a total of 5 immunizations. From the third immunization, blood was collected from the jugular-clavicular vein one week after each immunization and serum titers were detected.
[0087] Leukocytes were isolated from the peripheral blood after the fifth immunization, total RNA was extracted, and the VHH gene was cloned by reverse transcription PCR and nested PCR (wherein, the systems and parameters of reverse transcription PCR and nested PCR are described below)...
Embodiment 2
[0113] Example 2. Screening of SARS-CoV-2 Nanobodies
[0114] The first well of a 96-well microtiter plate was coated with the SARS-CoV-2 virus S protein antigen at a concentration of 1ug / mL, overnight at 4°C; the next day, the coating solution was poured out and washed three times with PBST. Block the first and second wells of the ELISA plate with BSA and incubate at room temperature for 2 hours; pour out the blocking solution and wash with PBST for 3 times; add the phage nanobody library obtained in Example 1 to the first well and react for 2 hours; Discharge the liquid, pat dry on clean absorbent paper, and wash 5 times with PBST; add 100 μL of S1 protein of SARS-CoV-2 virus Wild type original strain to the first well, and react for 1 h; aspirate the first well The liquid was added to the second well, reacted for 1 h, and the phage bound to BSA was removed; the eluate was collected, and 5 μL was used for titer determination, and the rest was used for amplification.
[0115...
Embodiment 3
[0117] Example 3. Expression of SARS-CoV-2 Nanobodies
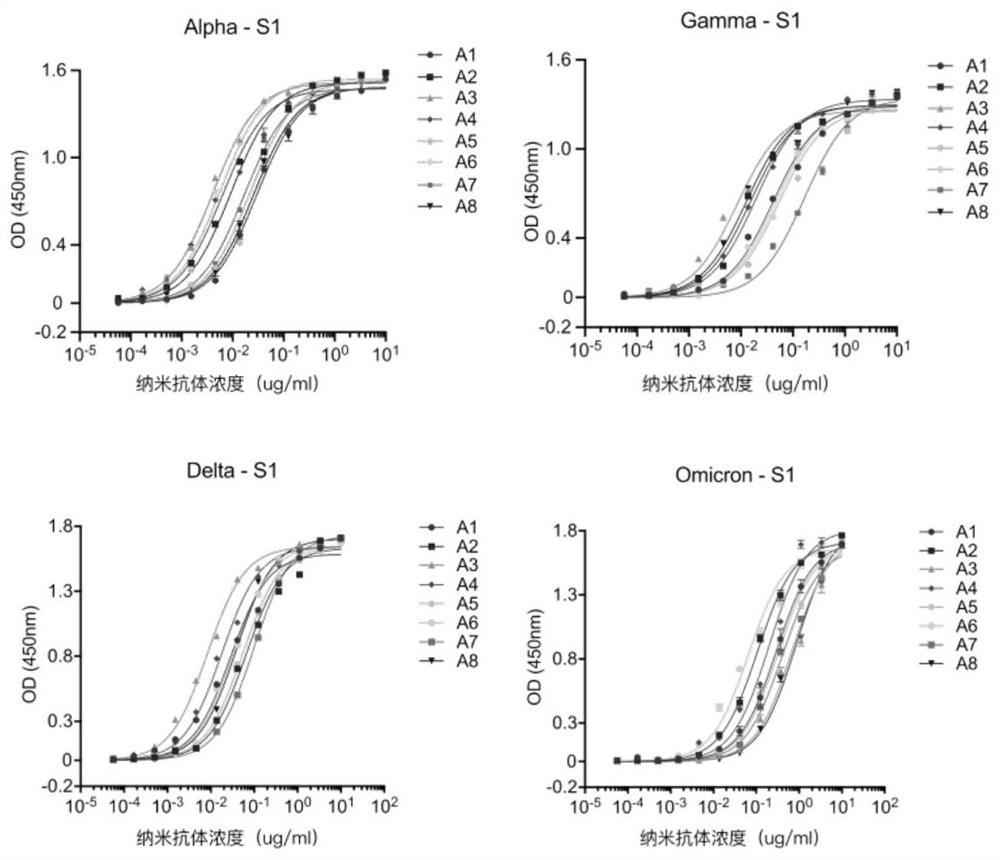
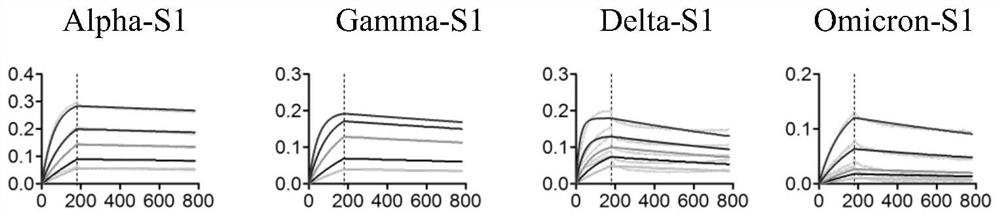
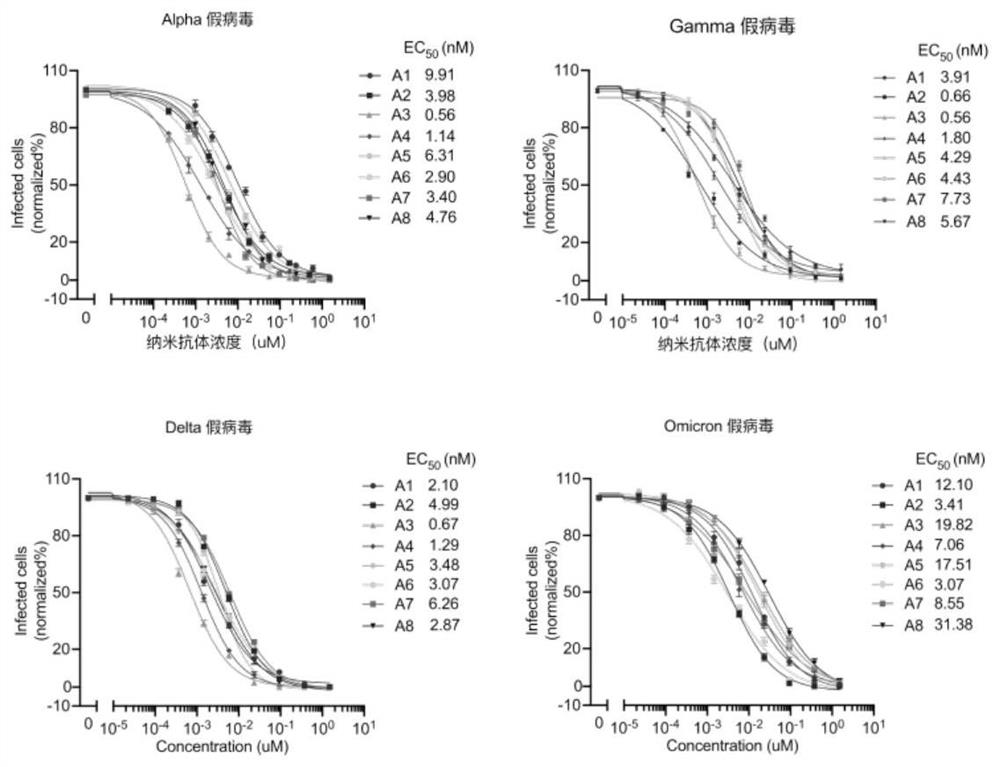
[0118] The positive monoclonal plasmid was extracted, transformed into E. coli TOP10F' competent cells (purchased from ThermoFishier), and then spread on solid medium for overnight culture after recovery. The next day, a single clone was picked and cultured in SB-carboxybenzyl medium, and IPTG was added to induce overnight expression; the next day, the cells were lysed with a high-pressure homogenizer, filtered through a membrane, and purified with a nickel column, that is, using a histidine tag to bind with the cells. The nanobodies were separated and purified by affinity chromatography of nickel chloride in the nickel column to obtain high-purity anti-SARS-CoV-2 nanobodies, namely antibodies A1-A8. After amino acid sequencing analysis, the amino acid sequences of the obtained nanobodies were as shown in SEQ IDNO: 1-8 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com