Mycobacterium vaccae formic dehydrogenase mutant and its use
A formate dehydrogenase and carrier technology, applied in the fields of application, bacteria, fungi, etc., can solve the problems of increased stability, increased copper stability, and decreased formate dehydrogenase activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0163] Subcloning of Formate Dehydrogenase from Mycobacterium vaccae
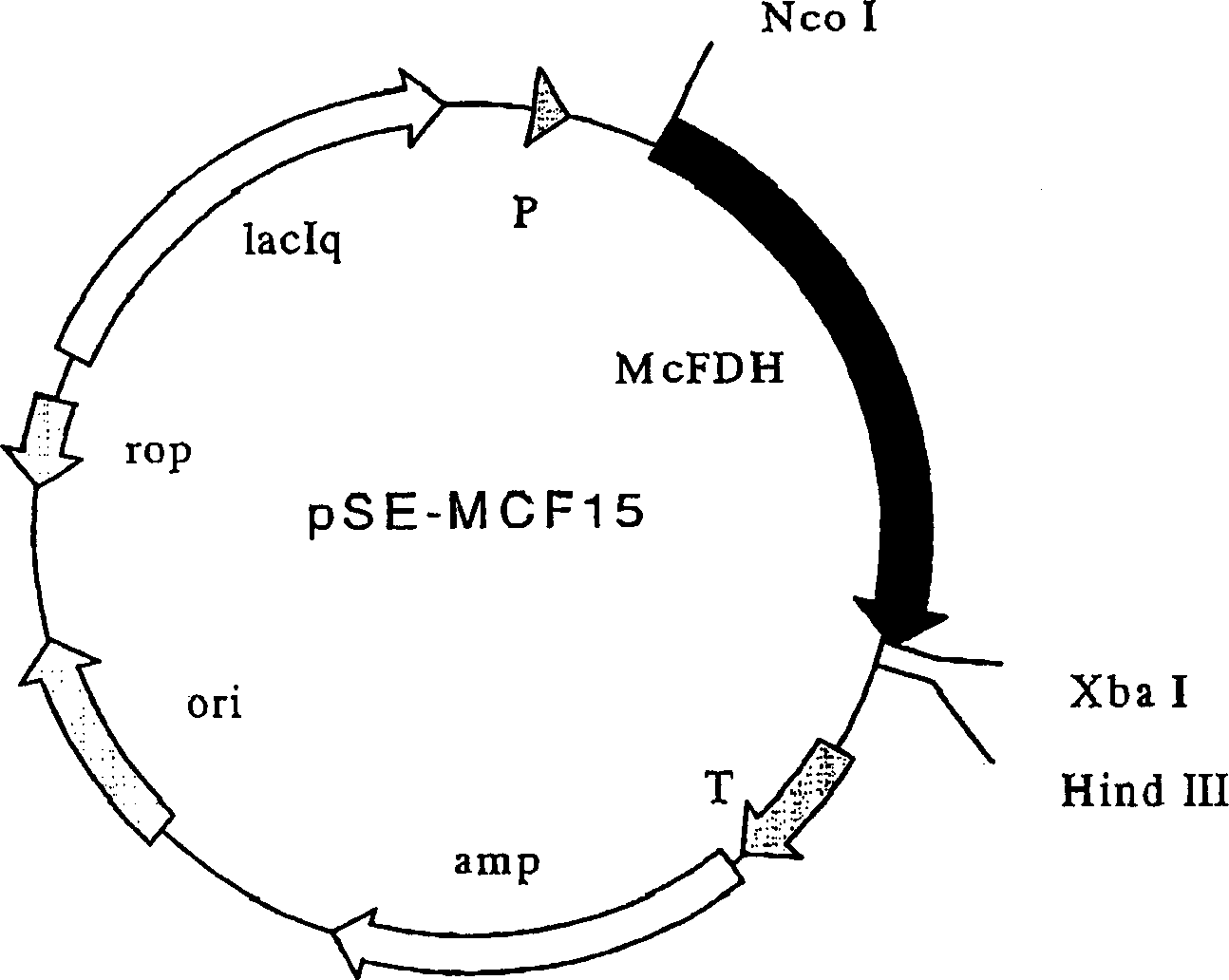
[0164] Formate dehydrogenase was subcloned from the plasmid pMcFDH(E-P) described in a reference (Appl. Microbiol. Biotechnol.), 44, 479-483 (1995). Primers MCF-ATG2 (5'-CTTTCTAGAGGAATTCAACCATGGCAAAAGTTCTGTGTGTTC-3' / SEQ ID NO: 3) and MCF-TAA3 (5'-CAGTCTAGATTAGACCGCTTTTTTGAATTTGGCG-3' / SEQ ID NO: 4) to clone the region containing only the open reading frame of formate dehydrogenase. The plasmid (pMcFDH(E-P)) was used as a template to carry out PCR and cycled 30 times (95°C, 45 seconds, 50°C, 1 minute, 75°C, 7 minutes) to obtain specific amplified DNA. The resulting DNA fragment was double-digested with two restriction endonucleases NcoI and XbaI. Plasmid vector pSE420D (JP-A 2000-189170) was double-digested with NcoI and XbaI, and the PCR-amplified DNA fragment double-digested with these enzymes was ligated thereto with T4 DNA ligase; pSE-MCF15 was thus obtained. The map of the plasmid is shown in the atta...
Embodiment 2
[0166] Construction of co-expression vector pSFR415 of formate dehydrogenase from Mycobacterium vaccae and carbonyl reductase from Kluyveromyces erylieii
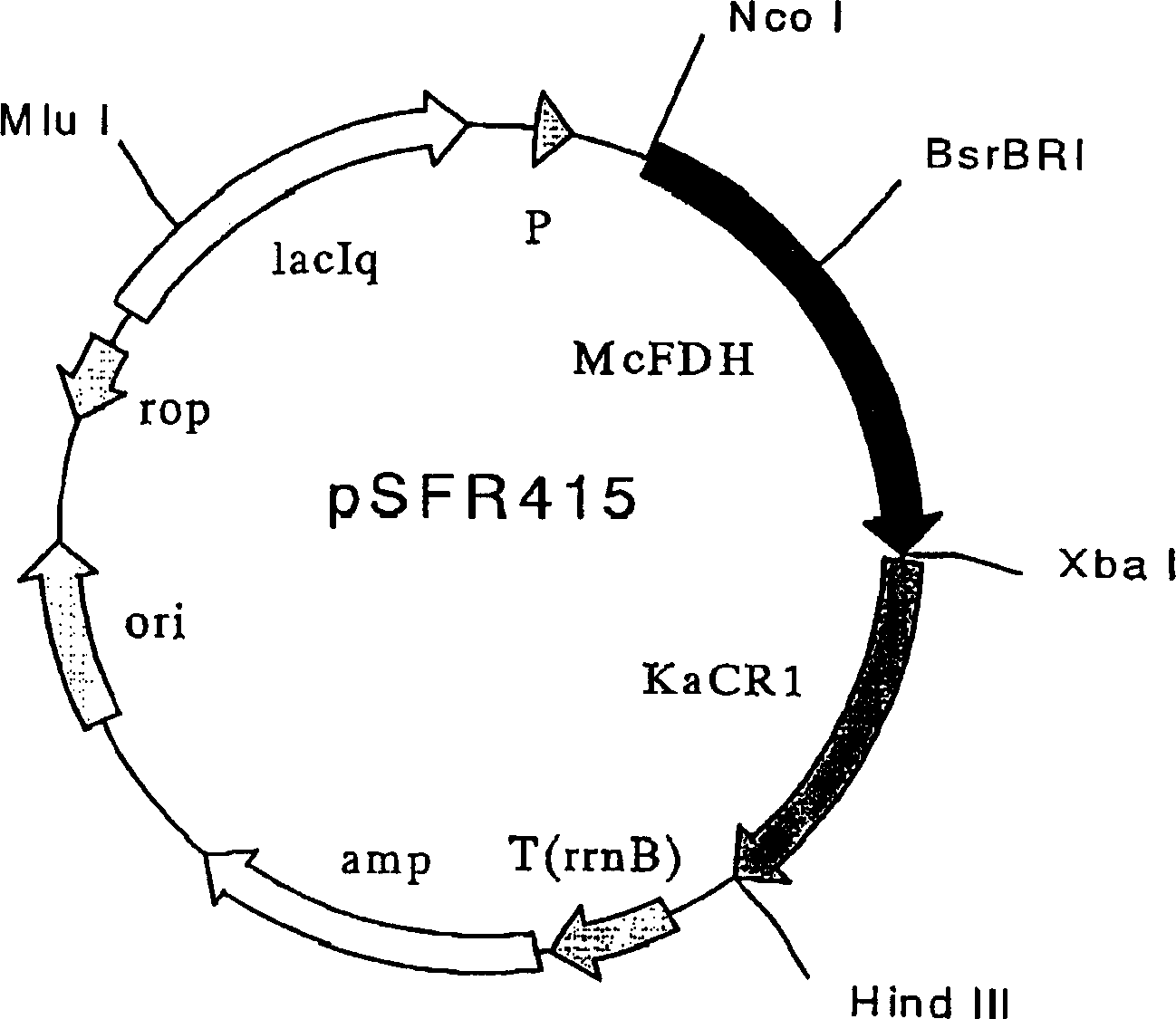
[0167] In order to clone the region containing only the open reading frame of carbonyl reductase derived from Kluyveromyces eleminia by PCR, primers KAR-BSG5-3 (5'-TAATCTAGAGGAATTCAATAATGGATCCAACAATGACGTTTC-3' / SEQ ID NO: 5) and KAR - BSG3 (5'-TAGAAGCTTAAGCTATTAAACGCAAGTGTACCCAC-3' / SEQ ID NO: 6). Using pSE-KAR1 (JP-A 2000-236883) as a template, PCR was performed 30 times (95°C, 30 seconds, 50°C, 1 minute, 75°C, 5 minutes) to obtain specifically amplified DNA. The resulting DNA fragment was double-digested with two restriction endonucleases XbaI and HindIII. The plasmid pSE-MCF15 containing the formate dehydrogenase derived from Mycobacterium vaccae constructed in Example 1 was double-digested with XbaI and HindIII two restriction endonucleases, and was combined with T4 DNA ligase A PCR-amplified DNA fragment digested with ...
Embodiment 3
[0176] Introduction of mutations by PCR into a formate dehydrogenase from Mycobacterium vaccae
[0177] On the basis of the co-expression vector pSFR415 derived from the formate dehydrogenase of Mycobacterium vaccae and the carbonyl reductase derived from Kluyveromyces eleminus constructed in Example 2, the Primer MCF-ATG3 (5'-CTTTCTAGAGGAATTCAACCATGGCAAAAGTTCTGTCTGTTC-3' / SEQ ID NO: 7) with serine replacing cysteine at the 6-position of Bacillus formate dehydrogenase and formate dehydrogenase 256 derived from Mycobacterium vaccae Primer McFDH-7mut01 (5'-GTATCCGGTTTGCGACGTCGTGACGCTGAACTCCCCGCTGCACCCCGAA-3' / SEQ ID NO: 8) and primer McFDH-7mut02 (5'-TTCGGGGTGCAGCGGGGAGTTCAGCGTCACGACGTCGCAAANOCCGGATAC-3' / SEQ ID) with cysteine replaced by serine at the position. Substitution of cysteine with serine at position 6 is hereinafter referred to as "C6S". According to this rule, the replacement of cysteine with serine at position 256 is called "C256S".
[0178] The first round o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com