Detection type gene chip for detecting various peptitis
A gene chip and hepatitis technology, which is applied in the field of gene chips for detecting hepatitis mutations, can solve the problems that it is difficult to replace conventional detection methods, the chip detection cost is high, and the detection cost is high, so as to facilitate large-scale promotion and application and reduce the amount of samples to be tested. The effect of less, high degree of automation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
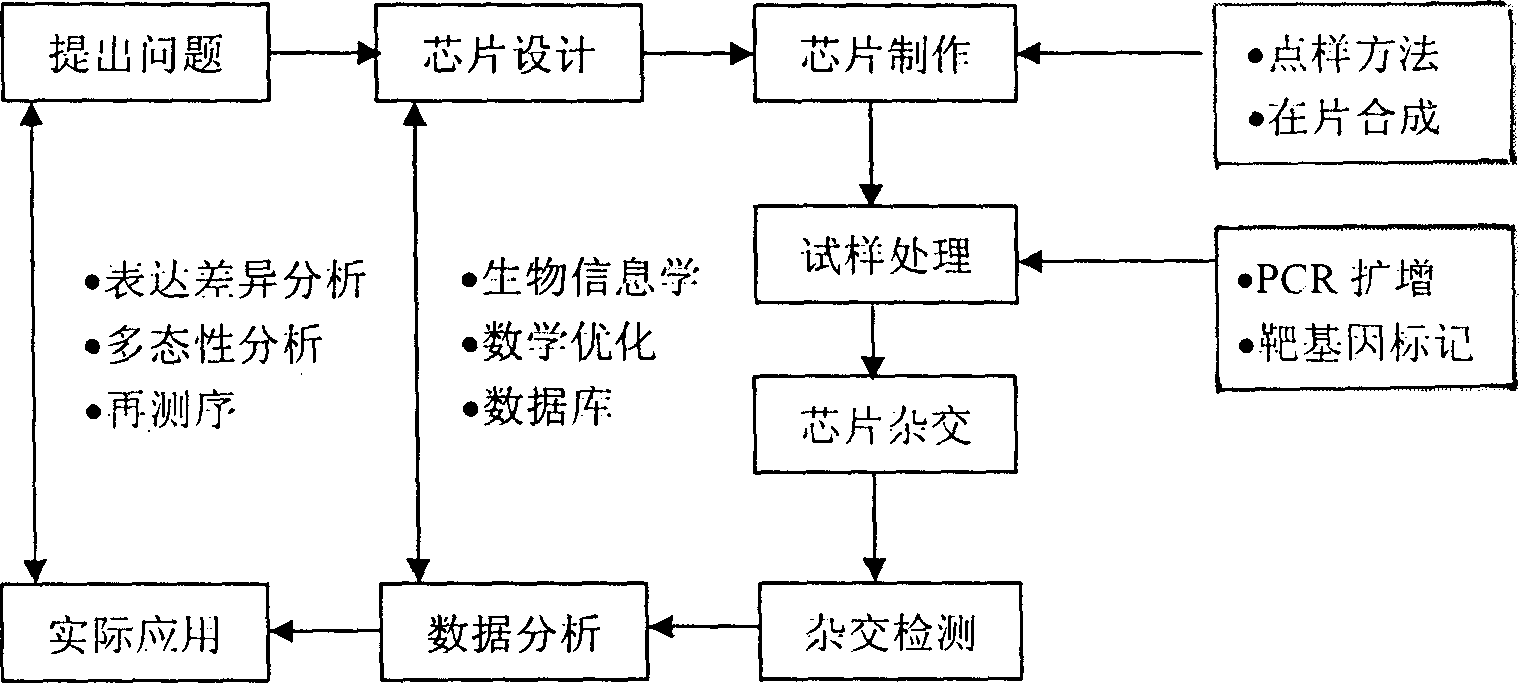
Image
Examples
Embodiment 1
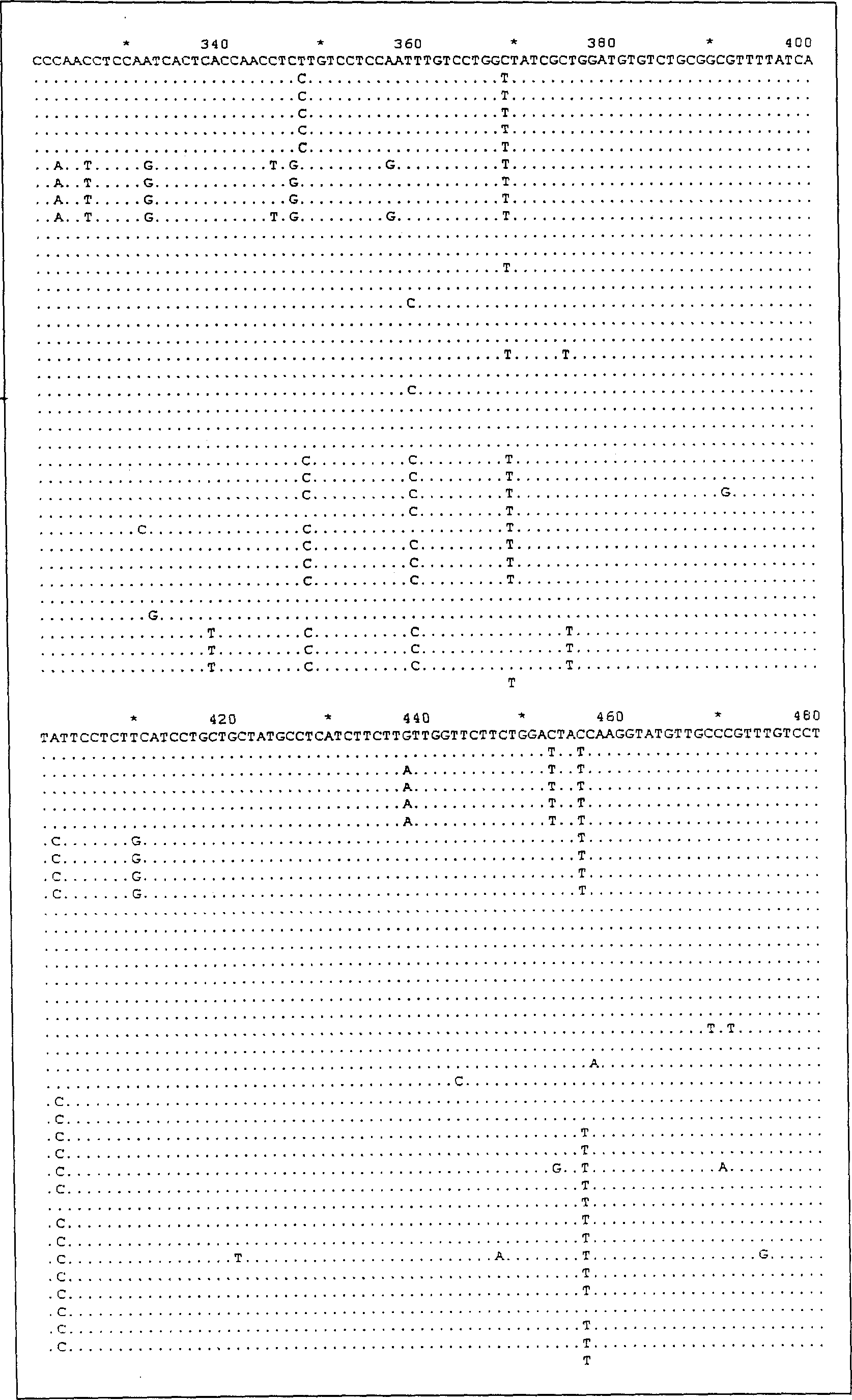
[0059] 1. Primer sequence:
[0060] Wherein the HBV primer1 sequence is as follows:
[0061] 5′-GTTTTATTCATATTCCCTCT-3′
[0062] 5′-GGTTTTTATTAGGGTTCAA-3′
[0063] The HBV primer2 sequence is as follows:
[0064] 5′-GAGGCTGTAGGCATAAAT-3′
[0065] 5′-GGGCATTTGGTGGTCTG-3′
[0066] A total of two pairs of primers were designed for HCV amplification primers,
[0067] Wherein the HCV primer1 sequence is as follows:
[0068] HCVi: 5′-GCCTTGTGGTACTGC-3′
[0069] HCVlC: 5'-5ACCGCTCGGAAGTC-3'
[0070] The sequence of HCV primer21 is as follows:
[0071] HCV2: 5′-GGCTTTACCGGCGAC-3′
[0072] HCV2C: 5′-GCTCATACCAiGCAC-3′
[0073] 2. DNA and RNA extraction from serum samples
[0074] Add 100 μl of serum to 10 μl of DEPC-ethanol solution (10% diethylpyrocarbonate (DEPC), 90% absolute ethanol) and let it stand at room temperature for 10 minutes, then in a boiling water bath for 10 minutes, then dry the tube cover at 65°C for 15 minutes. Minutes, centrifuged for 5 minutes (12000 rpm...
Embodiment 2
58.0
CCACACGTCG CGCCTCATTT
[0095] 2. Synthesize the above probe and label the amino group at the 5' end, dilute it with spotting buffer, spot the sample on the glass slide with the modified arm molecule with a spotting instrument, and fix it.
[0096] Serum sample DNA, RNA extraction
[0097] Add 100 μl of serum to 10 μl of DEPC-ethanol solution (10% diethylpyrocarbonate (DEPC), 90% absolute ethanol) and let it stand at room temperature for 10 minutes, then in a boiling water bath for 10 minutes, then dry the tube cover at 65°C for 15 minutes , centrifuged for 5 minutes (12000 rpm, centrifuged for 10 minutes), set aside.
[0098] 3. Prepare template DNA by reverse transcription
[0099] Add the following reagents in sequence to the eppendorf tube containing the DNA and RNA precipitates prepared in step 2:
[0100] Milli-Q H2O (ultrapure water) 11 μl, 15 μmol / L HCV, HCV, reverse transcription primer 2 μl and fully dissolve the precipitate, mix the sample, cen...
Embodiment 3
[0115] 1. Design probes for virus detection of HBV, HCV, HDV, HGV, TTV and other viruses; design probes for detection of subtypes of HBV and HCV; design the following probes for detection of mutation types of HBV viruses .
[0116] 2. Synthesize the above probe and label the amino group at the 5' end, dilute it with spotting buffer, spot the sample on the glass slide with the modified arm molecule with a spotting instrument, and fix it.
[0117] Serum sample DNA, RNA extraction
[0118] Add 100 μl of serum to 10 μl of DEPC-ethanol solution (10% diethylpyrocarbonate (DEPC), 90% absolute ethanol) and let it stand at room temperature for 10 minutes, then in a boiling water bath for 10 minutes, then dry the tube cover at 65°C for 15 minutes. Minutes, centrifuged for 5 minutes (12000 rpm, centrifuged for 10 minutes), set aside.
[0119] 3. Prepare template DNA by reverse transcription
[0120] Add the following reagents in sequence to the eppendorf tube containing the DNA and RN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com