High efficiency transglyco beta galactoside gene
A technology of galactosidase and transglycosylation, which is applied in the directions of genetic engineering, plant gene improvement, enzymes, etc., can solve the problem that there is no Enterobacter cloacae transglycosyl β-galactosidase gene report, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
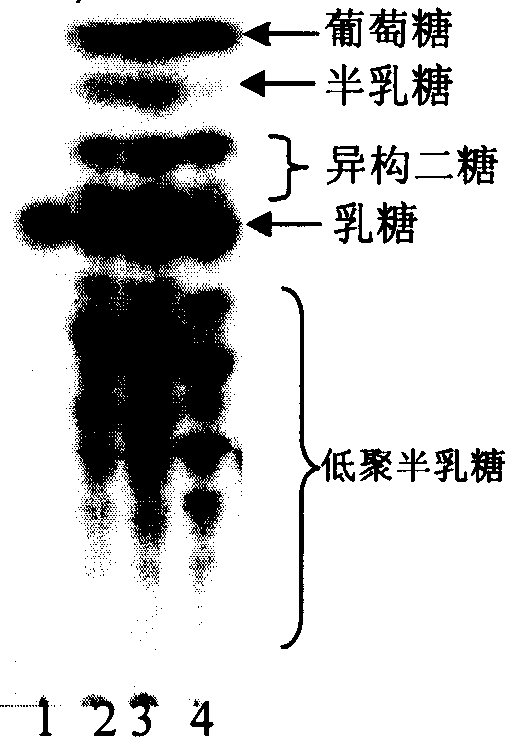
Image
Examples
Embodiment 1
[0025] Example 1 Extraction of Enterobacter cloacae B5 Genomic DNA
[0026]Inoculate Enterobacter cloacae B5 CGMCC No.1401 in 5 mL of LB medium, culture on a shaker at 37°C for 16 hours, take 1.5 mL of the culture, centrifuge at 12,000 rpm for 2 minutes, and suspend the obtained pellet in 565 μL of TE buffer solution; add 30 μL 10% (mass volume ratio) sodium dodecyl sulfate (SDS) and 5 μL 20 mg / mL proteinase K, mix well, and incubate at 37°C for 1 hour; add 100 μL 5mol / L NaCl and mix well, then Add 80 μL CTAB / NaCl solution, mix well, and incubate at 65°C for 20 minutes; then ice-bath for 30 minutes; add an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1, v / v / v), mix Centrifuge at 12,000 rpm for 5 minutes, transfer the supernatant to a new tube, add an equal volume of phenol / chloroform / isoamyl alcohol, and mix well; centrifuge at 12,000 rpm for 5 minutes, transfer the supernatant to a new tube Add 2 times the volume of absolute ethanol to the tube, mix gently until ...
Embodiment 2
[0030] Example 2 PCR amplification of E.cloacae B5β-galactosidase gene
[0031] Design primers to introduce the BamHI restriction site that can be inserted into the pET-15b plasmid (Novagen) (the pET-15b plasmid vector has an N-terminal 6His tag, which is convenient for the purification of expressed proteins), and the sequence is as follows:
[0032] F-B5: 5'-GATCGGATCCGATGCCCAACACTCTATCG-3'
[0033] R-B5: 5'-GAAGGGATCCTTAAGGGTTCTGCTGCCA-3'
[0034] Using the extracted Enterobacter cloacae genomic DNA as a template, the above primers were used for PCR amplification. The total volume of the PCR amplification system is 50 μL, containing 35 μL of ultrapure water, 5 μL of 10×PCR buffer, dNTP 2.4 mmol / L, primers 1 μmol / L, MgCl 2 1.5mmol / L, template DNA 100ng, TaKaRa LA Taq 2.5U. PCR amplification conditions: pre-denaturation at 95°C for 5 minutes; reaction for 28 cycles (denaturation at 95°C for 1 minute, annealing at 52°C for 1 minute, extension at 72°C for 3 minutes); extensio...
Embodiment 3
[0035] Cloning and DNA sequence determination of embodiment 3 PCR products
[0036] The PCR recovery product of embodiment 2 is connected on the pMD18-T plasmid carrier (TA clone), transforms Escherichia coli Top10 competent cell, spreads ampicillin plate, screens recombinant bacteria with colony PCR method, then sequenced, sequenced by Shanghai Bo Subbiotech Co., Ltd. completed the analysis of the measured sequence, showing that there is an open reading frame consisting of 3090 bases, encoding 1029 amino acids, which is the complete gene sequence of E.cloacae B5β-galactosidase, such as SEQ ID Shown in No.1.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Km value | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com