Method for screening excellent breeding cattle with normal CD18 gene and dedicated primer therefor
A technology for normal and breeding cattle, applied in biochemical equipment and methods, recombinant DNA technology, and microbial assay/inspection, etc. Promising effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1. Primer design and optimization of PGR reaction conditions for screening good breed cattle with normal CD18 gene
[0028] 1. Primer design
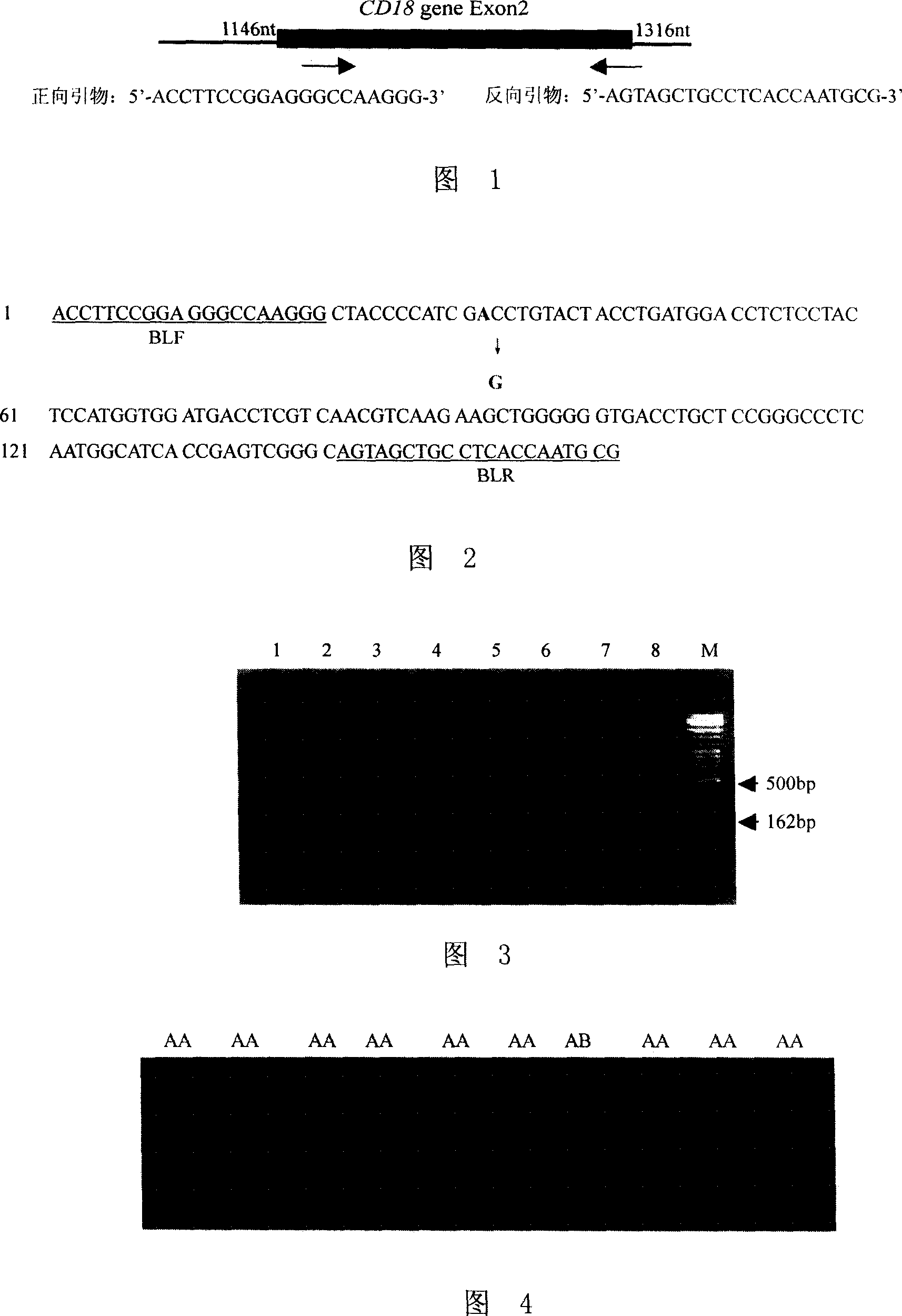
[0029] According to the nucleotide sequence of the bovine CD18 gene (GenBank number: Y12672), use Oligo6.0 software to design specific PCR primers for PCR-SSCP screening of fine breed cattle with normal CD18 gene, and the length of the PCR product is about 150-300bp for comparison Suitable for PCR-SSCP detection. Through the design, screening and optimization of a series of PCR primers, finally according to the nucleotide sequence shown in sequence 3 in the sequence listing, the sequence contains the mutation site that causes BLAD (the 32nd base, namely A55G:CD18 From the 55th base at the 5' end of the gene Exon2, which is base A for normal cattle and base G for BLAD cattle), a primer BLF (upstream primer) is designed: 5'-ACCTTCCGGAGGGCCAAGGG-3' (sequence 1 in the sequence listing) And primer BLR (downstream primer): 5'...
Embodiment 2
[0032] Example 2, PCR-SSCP Screening of Excellent Breed Cattle with Normal CD18 Gene and Its Sequencing Identification
[0033] 1. PCR-SSCP screening
[0034] Extract the genomic DNA of the fresh blood of 54 bulls and use it as a template, and carry out PCR amplification under the guidance of the specific primer pair BLF / BLR obtained in Example 1. The PCR reaction system is the same as in Example 1, and the reaction conditions are: Pre-denaturation at 94°C for 5min; denaturation at 94°C for 30s, annealing at 64°C for 30s, extension at 72°C for 30s, and 35 cycles; extension at 72°C for 7min. After the reaction, 1.5 μl of the PCR amplification products of the 54 bulls were placed in PCR tubes, and 6 μl of loading buffer (98% formamide, 10 mM EDTA pH8.0, 10% glycerol, 0.025% bromophenol blue and 0.025 xylene cyanine), after denaturing at 98°C for 10 minutes, immediately place it on ice for 5 minutes, apply samples, and perform 14% non-denaturing polyacrylamide gel electrophoresi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com