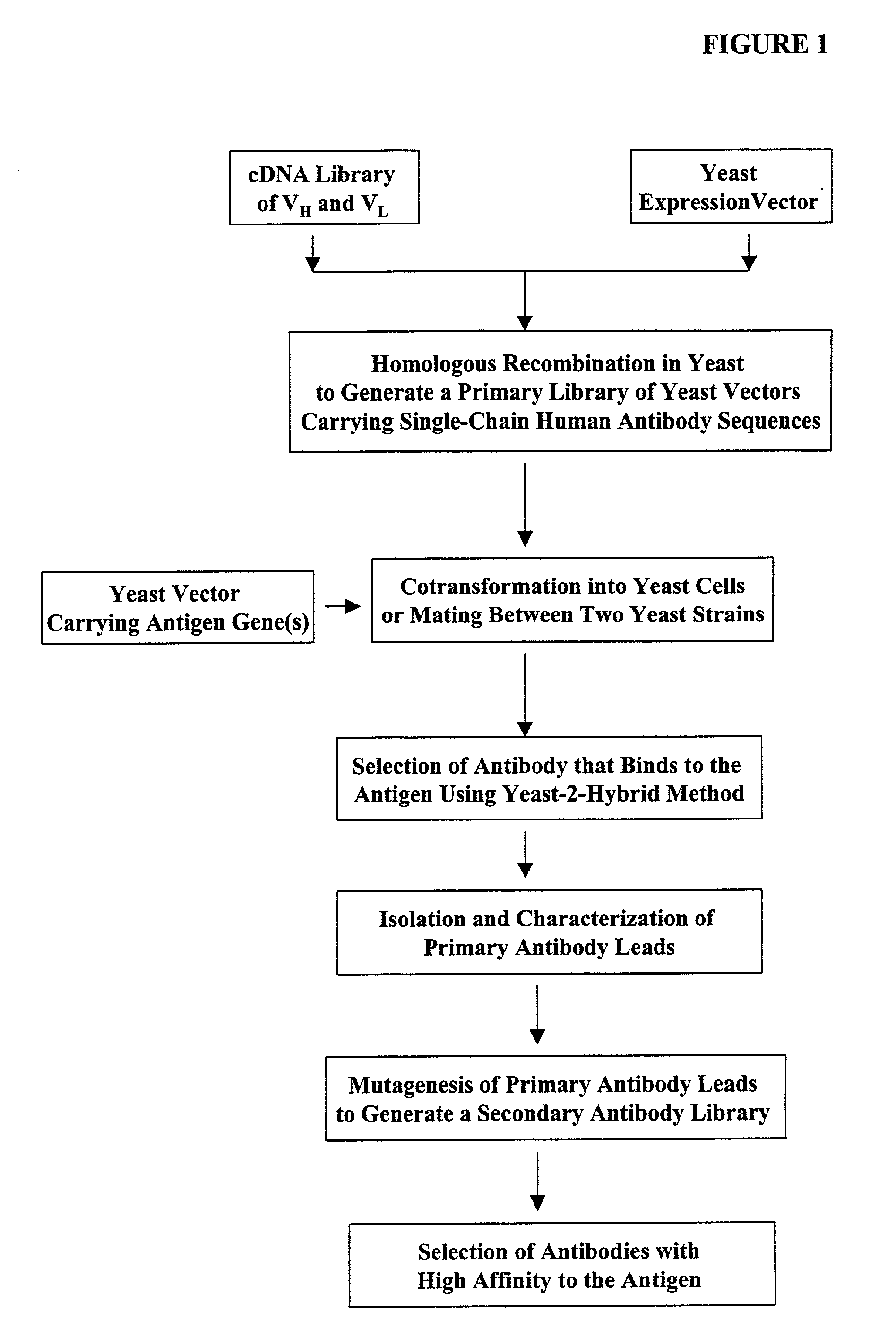
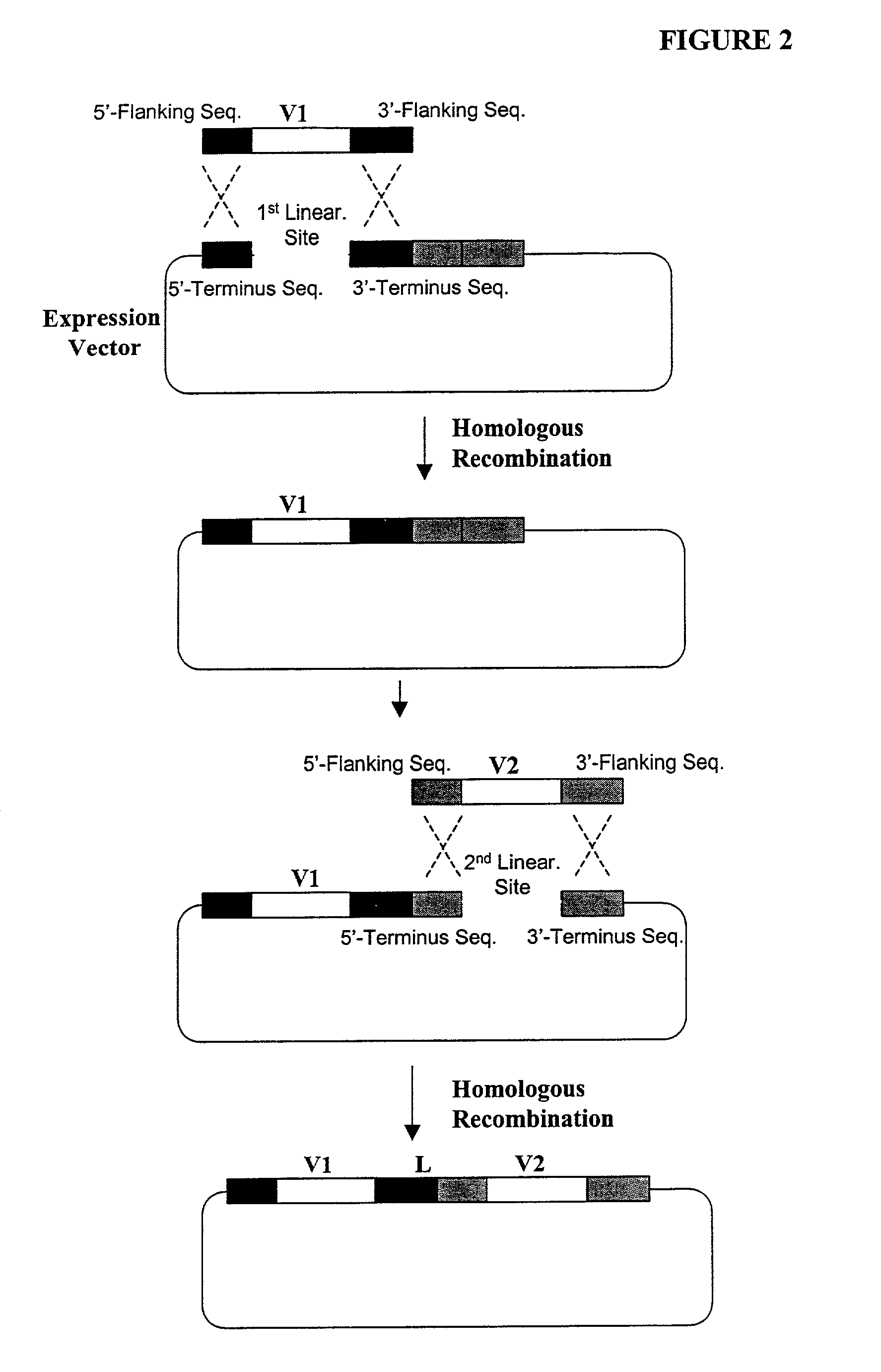
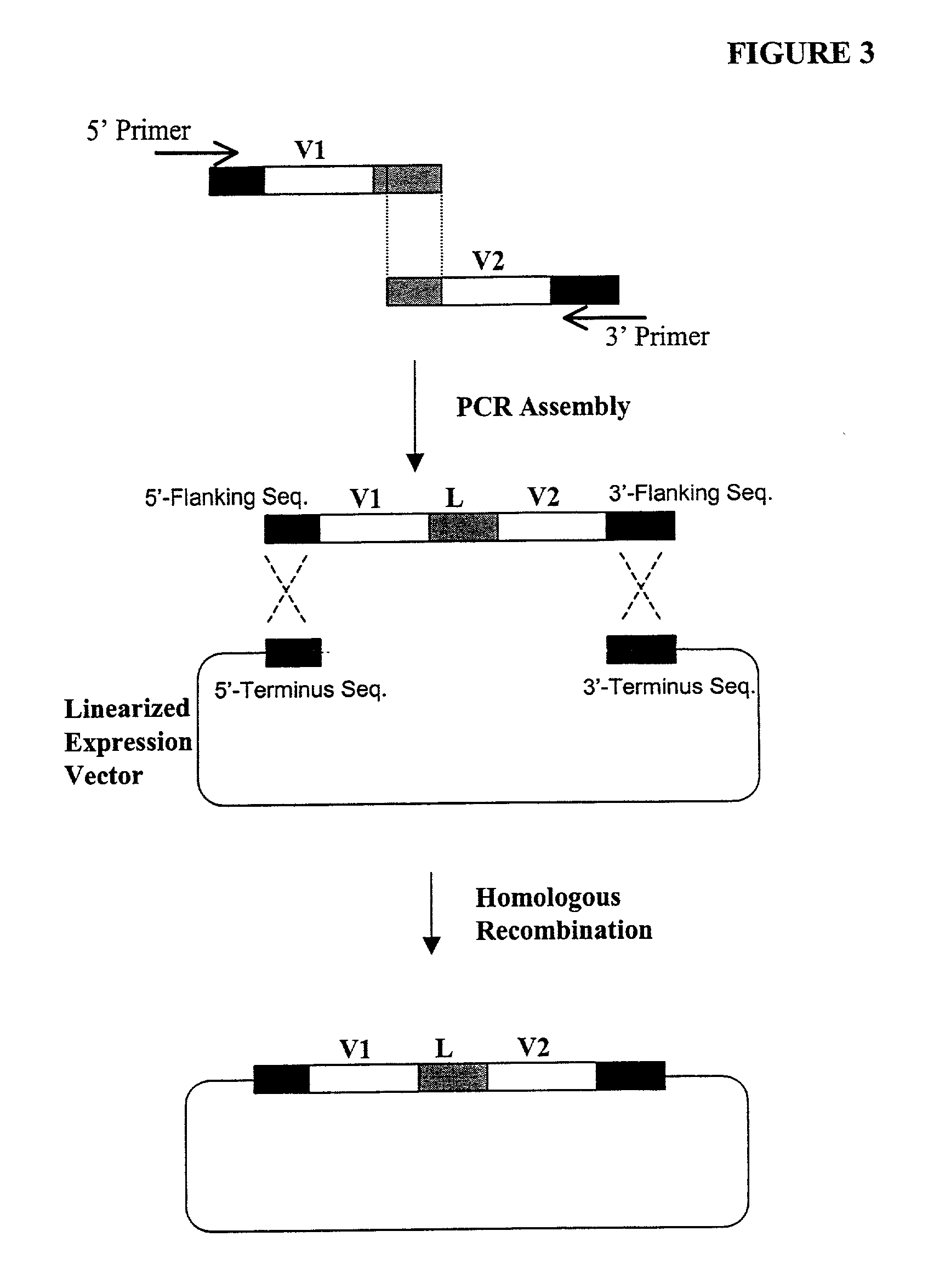
Generation of highly diverse library of expression vectors via homologous recombination in yeast
a technology of expression vectors and homologous recombination, which is applied in the field of generating libraries of recombinant expression vectors, can solve the problems of long selection period of high affinity antibodies, bacteria that cannot readily process, assemble, express/secrete functional antibodies,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Expression Vectors Containing Human Single-Chain Antibody scFv Library Using Homologous Recombination In Vivo
[0380] The following illustrates examples of how to use general homologous recombination as an efficient way of constructing recombinant human scFv library. The coding sequence of each member of the scFV library includes a heavy-chain variable region V.sub.H and a light-chain variable region V.sub.L derived from a library of human antibody repertoire. The scFv library is fused with a two-hybrid system activation domain (AD) to form a two-hybrid expression vector in the yeast.
[0381] 1) Isolation of Human scFv cDNA Gene Pool
[0382] A complex human scFv cDNA gene pool is generated by using the method described in Sambrook, J., et al. (1989) Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.; and Ausubel, F. M. et al. (1995) Current Protocols in Molecular Biology" John Wiley & Sons, NY.
[0383] Briefly, total RNA is isolat...
example 2
Construction of Human scFv library by Using CRE / loxP-Mediated Recombination in Vivo
[0413] In this example, the construction of a highly complex and diverse combinatorial repertoire in yeast using V-region gene segments as building blocks is described.
[0414] First, a special type of human scFv library is generated in yeast by the standard homologous recombination procedure underlined in Example 1. This library is consisting of 10.sup.7 or more of highly diverse and complex V-region gene repertoire derived from heavy chain and light chain origin. One pool (e.g., VL or light chain gene segment) is flanked on both sides by two non-identical lox P sites. The loxP sites are designed into the primer sequences used in one of the PCR amplification steps. Examples of the loxP sites are listed in Table 1.
[0415] Specifically, two nonidentical loxP sites, loxP1 [SEQ ID NO: 4] and loxP2 [SEQ ID NO: 5] (Table 1), are incorporated into the PCR primers for amplifying the V.sub.H and V.sub.L gene seg...
example 3
Construction of Human scFv Library of Very High Complexity by Using CRE / loxP-Mediated Recombination in Vivo-Second Design
[0432] An alternative method to the method described in Example 2 for construction of human scFv library using CRE / loxP-mediated recombination is to use a "forced" multiple transformation. In this design, two starting human scFv libraries containing human heavy and light chain gene segments are generated separately in two vectors with different selection markers (e.g., Leu 2 and Ade 2, respectively). By selection of both markers will ensure that every yeast cell have both types of library clones (each may have multiple but variable number of copies). The activation or expression of Cre combinase in the yeast should allow the CRE / loxP-mediated recombination as illustrated in FIG. 4B.
[0433] Two special human scFv libraries are generated in yeast via homologous recombination by using the procedures described in Example 2. The two libraries are otherwise the same in t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| dissociation constant | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com