Methods and compositions for isolating small RNA molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0121] Preparation of Cell Lysate and Isolation of RNA
[0122] The following procedure provides the basis for the invention and is referred to in the Examples as the Ambion miRNA Isolation Kit (AMIK) procedure.
[0123] Frozen tissue was ground under liquid nitrogen to a fine powder. Lysis buffer (4 M GuSCN; 0.1 M beta-mercaptoethanol; 0.5% N-lauroyl sarcosine; 25 mM Na-citrate, pH 7.2) was added to this powder in an appropriate vessel at a proportion of 1 ml to every gram of tissue powder. This was homogenized using mechanical means to create a finely-dispersed tissue lysate. One tenth volume of a 2 M Na acetate (pH 4.0) solution was added and mixed thoroughly, adding 0.1 ml for every ml. The lysate was then processed immediately (without organic extraction) or placed on ice to be processed within 15 minutes.
[0124] Processing involved the addition of an equal volume of Acid Phenol-Chloroform (5:1, equilibrated with aqueous solution at pH 4.5) to the suspension, followed by vigorous a...
example 2
[0127] Detection of miRNAs through Northern Blotting
[0128] For each RNA sample, 5 μl was combined with 5 μl of Gel Loading Dye II (Ambion). Prior to loading on a denaturing acrylamide gel, these samples were heated at 95° C. for 2-5 minutes. The standard gel was 15% acrylamide (monomer:bis ratio of 19:1), 7M urea, buffered with TBE (Tris-Borate-EDTA, Peacock and Dingman, 1967).
[0129] The gel was routinely pre-run at 300-450 V for 30 minutes prior to loading the samples in sample buffer, which also contained bromphenol blue and xylene cyanol tracking dyes. The electrophoresis was performed at 300-450 V for 45-60 min, or until the bromphenol blue tracking dye was in the lower quadrant of the gel.
[0130] After electrophoresis, the gel apparatus was disassembled and the gel was electroblotted to a BrightStar-Plus Nylon membrane (Ambion). This procedure can be performed in a semi-dry apparatus using a stack of three sheets of Whatman filter paper (3MM) soaked in 0.25×TBE above and belo...
example 3
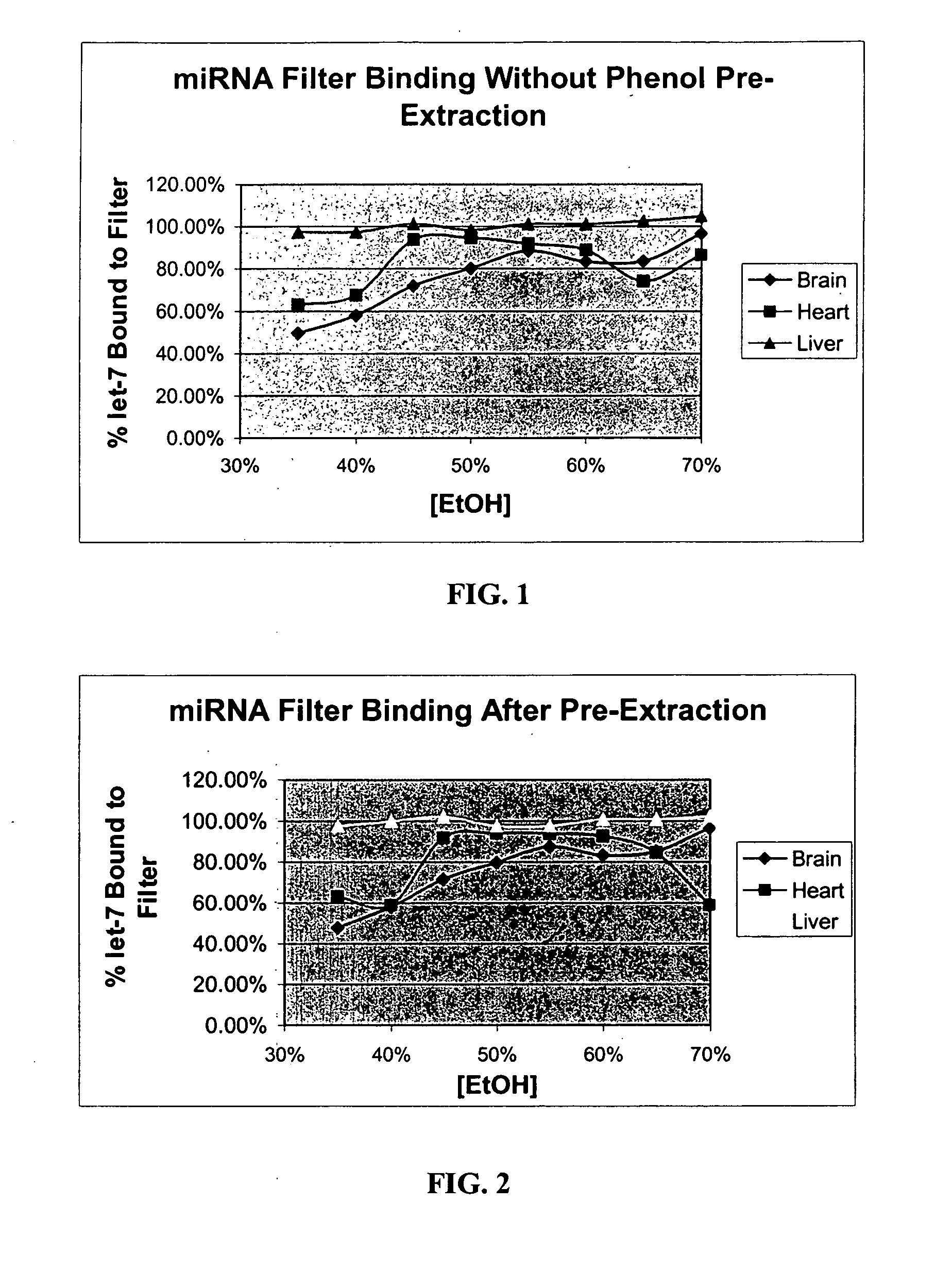
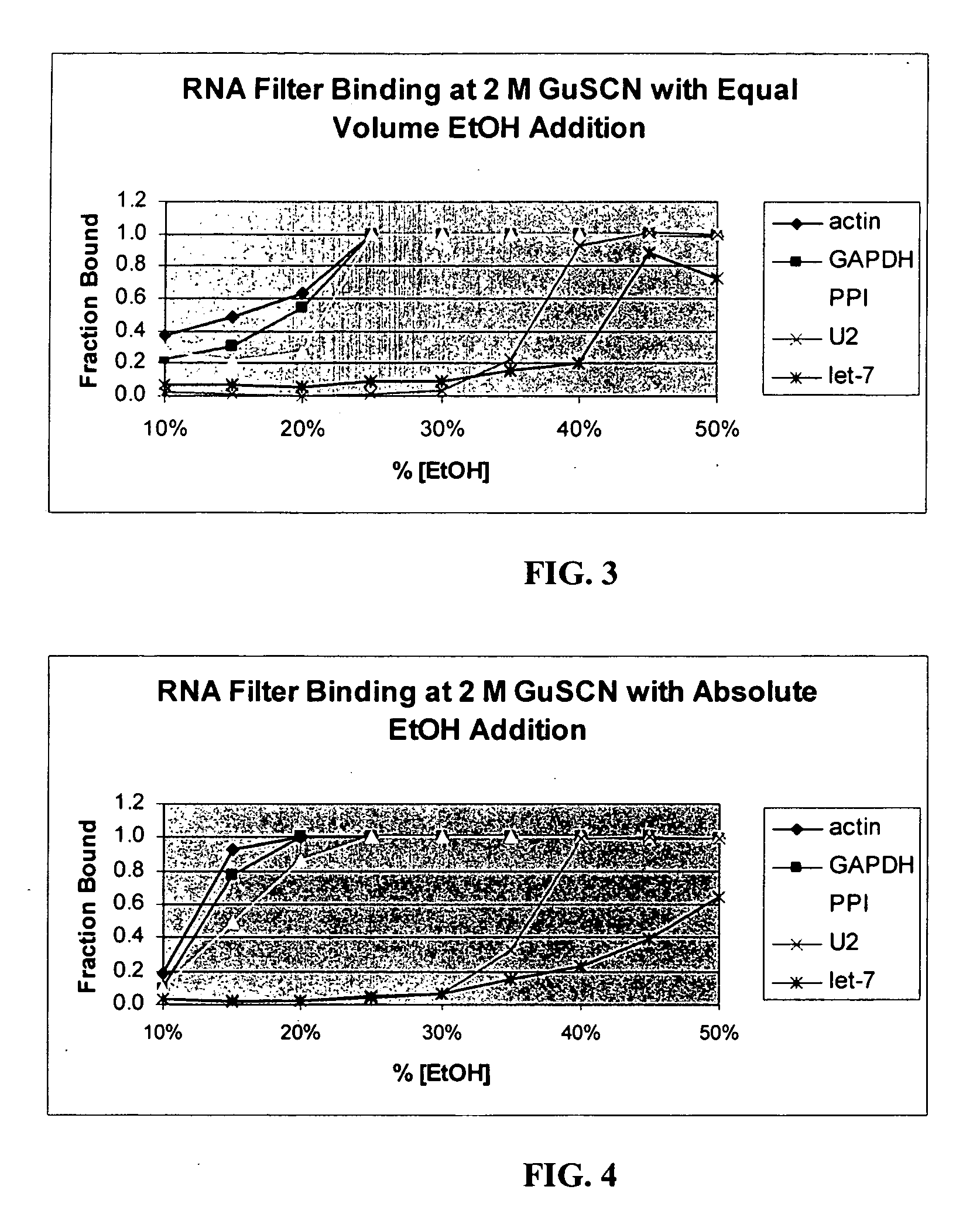
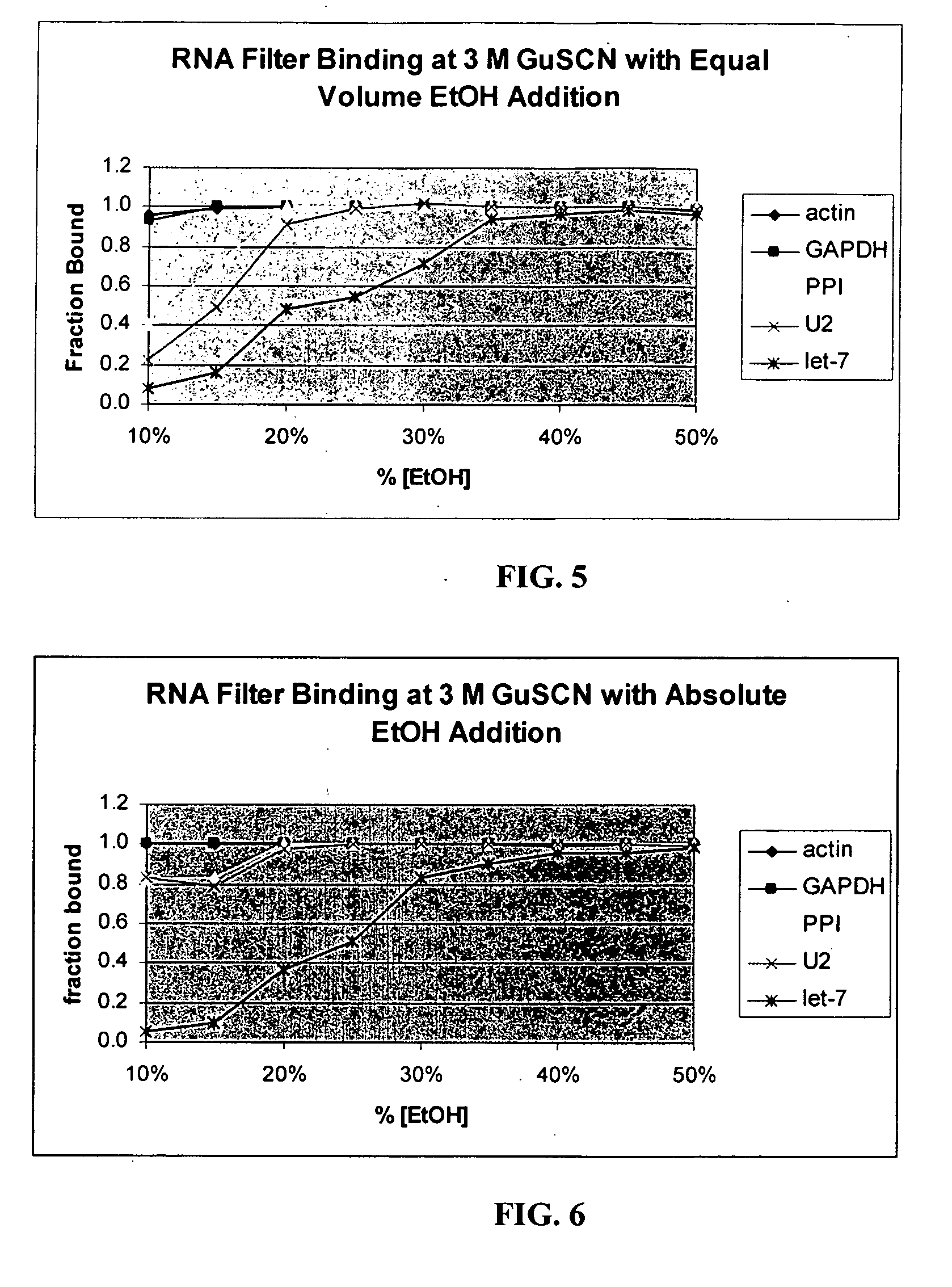
[0134] Enrichment of Small RNA Molecules
[0135] Frozen mouse brain, heart, liver, and kidney were processed separately according to the following protocol for enrichment of small RNAs.
[0136] Approximately one-half gram of frozen mouse (strain Swiss-Webster, 6-12 weeks old) tissue was crushed to fine powder under liquid nitrogen in a mortar. This powder was further dispersed in standard lysis buffer (4 M GuSCN; 0.1 M beta-mercaptoethanol; 0.5% N-lauroyl sarcosine; 25 mM Na-citrate, pH 7.2) by the use of a rotor-stator homogenizer with a 7 mm generator at high speed for ˜30 sec.
[0137] After homogenization, 0.6 ml of the lysate was removed for this study. 60 μl of 2M Na-acetate, pH 4.0, was added to the lysate, followed immediately by 0.6 ml of acid phenol-chloroform. After 30 sec of vigorous agitation, the aqueous phase was separated by centrifugation at 16,000×G for 5 min. Four 100 μl aliquots of this aqueous phase were used in four separate separations. The four aliquots had 100 μ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com