Flavobacterium heparinum expression system
a technology of flavobacterium heparinum and host system, which is applied in the field of new procaryotic expression system of flavobacterium heparinum, can solve the problems of inability to secrete recombinantly produced proteins, inability to glycosylate recombinantly produced proteins, and difficulty in purifying recombinantly produced proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
F. heparinum Plasmid Systems
[0028] PBBRICm is a derivative of the plasmid pBBR1 with a gene coding for chloramphenicol resistance cloned in. It is a cryptic plasmid isolated from Bordetella bronchiseptica S87. It is further a broad-host-range plasmid known to replicate in E. coli.
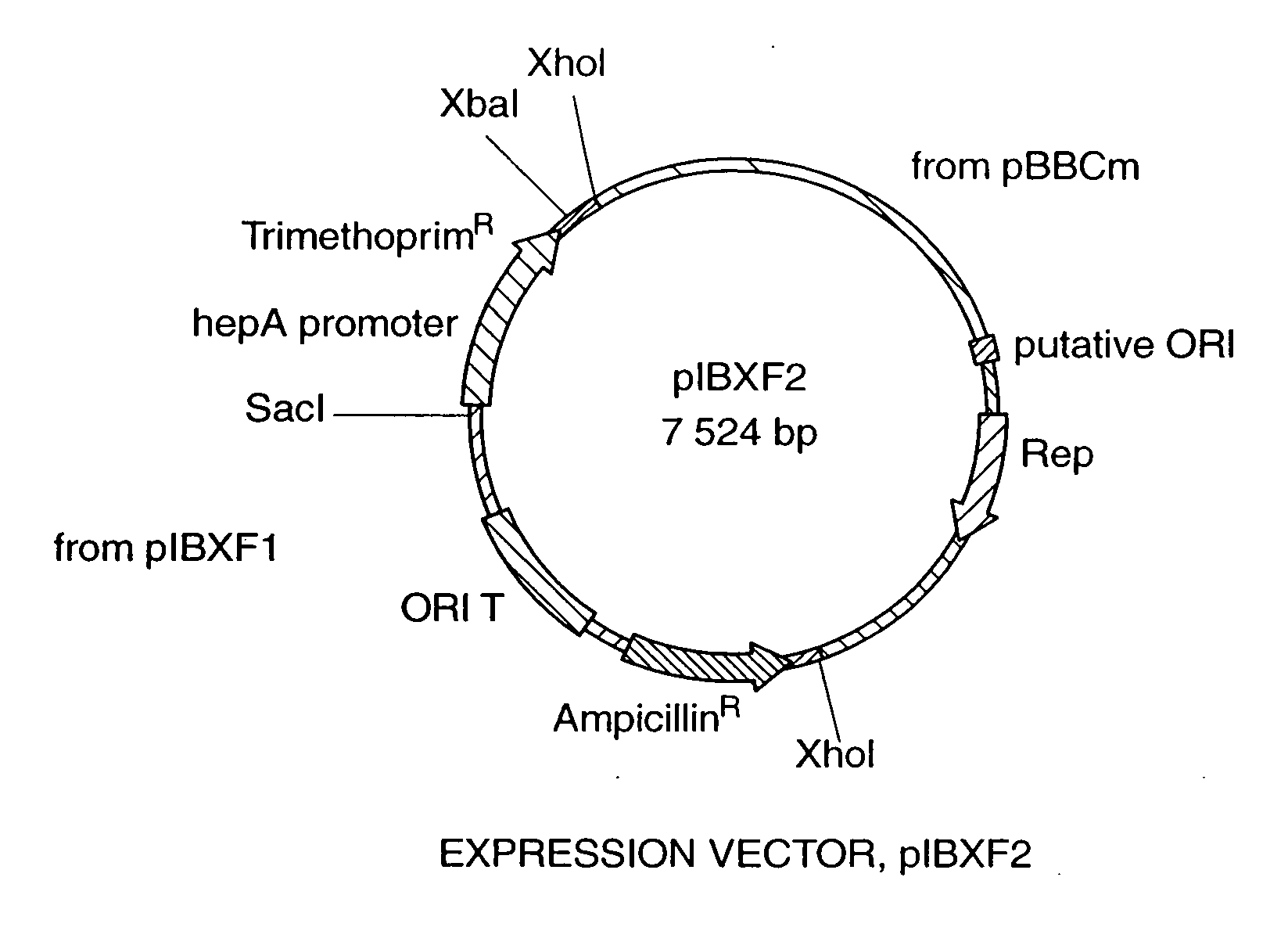
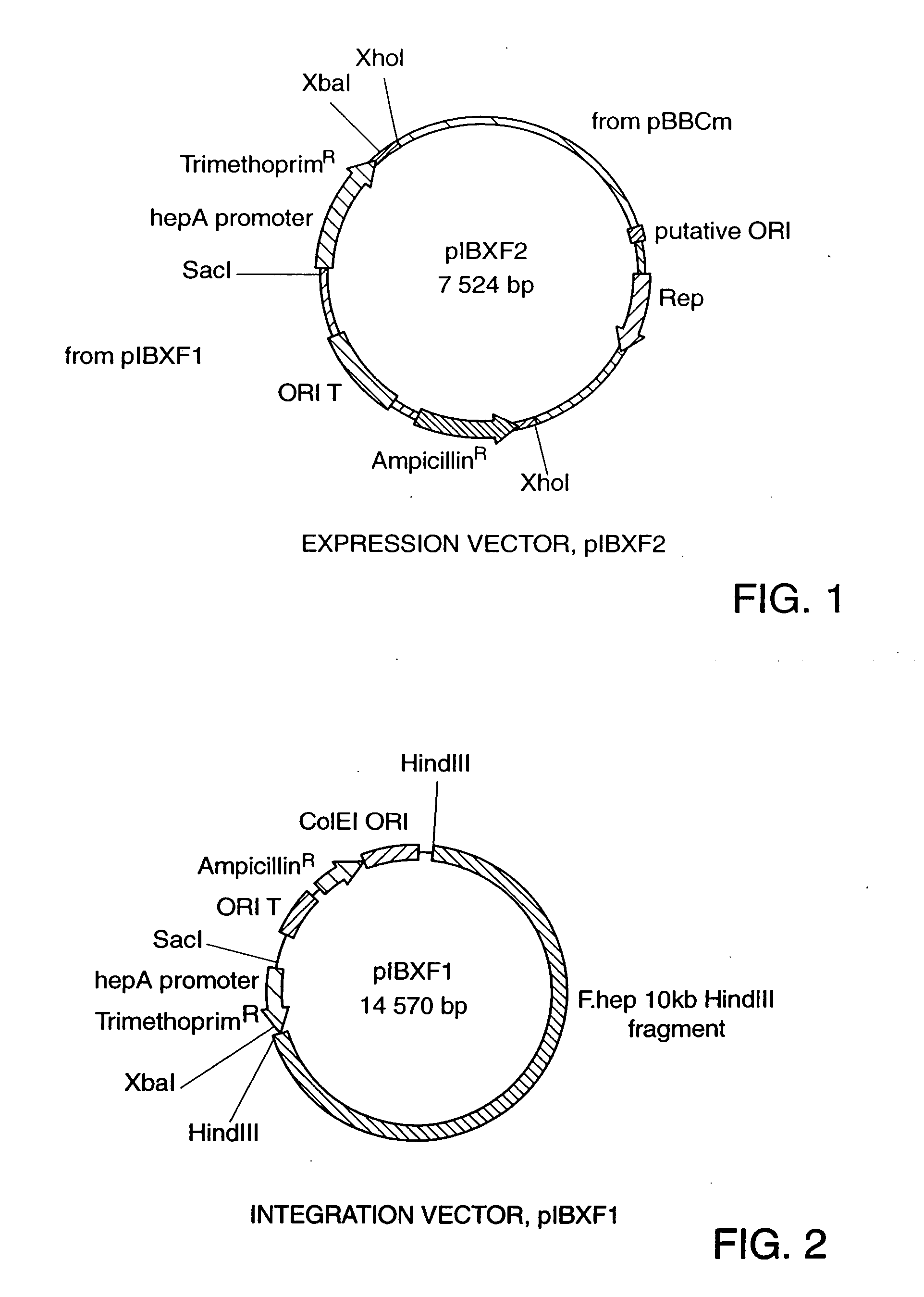
[0029] PIBXF2 has the following characteristics: [0030] (1) a bla gene coding for β lactamase (conferring ampicillin resistance)—functional in E. coli; [0031] (2) an ORIT region—allows for the mobilization of the plasmid into a new host (a Sau3 AI 307 bp fragment isolated from plasmid RP4); [0032] (3) a dhfrII gene coding for dihydrofolate reductase type II (conferring trimethoprim resistance)—cloned from plasmid R751 and placed under the control of the hepA promoter—functional in F. heparinum; [0033] (4) a hepA promoter—an 830 bp fragment 5′ of the hepA gene—induced by heparin; [0034] (5) a unique restriction site (for example, a XbaI site)—site for insertion of all homologous and heterologous genes; [00...
example 2
F. heparinum Integration Vector
[0040] The integration vector, pIBXF1, was created by addition of various elements to the parent plasmid, pUC13T.
[0041] The following elements are present on the parent plasmid: [0042] (1) bla gene coding for β lactamase (conferring ampicillin resistance)—functional in E. coli. [0043] (2) ColEI ORI—origin of replication—functional in E. coli. [0044] (3) ORIT region—allows for the mobilization of the plasmid into a new host (a Sau3 AI 307 bp fragment isolated from plasmid RP4).
[0045] The following elements were introduced into the pUC13T plasmid to confer integrative and selective functions in F. heparinum. [0046] (1) 10 kb HindIII fragment—a 10 kb fragment random selected from genomic F. heparinum DNA which was digested with HindIII, and cloned into pUC13T. [0047] (2) dhfrrII gene coding for dihydrofolate reductase type II (conferring trimethoprim resistance)—cloned from plasmid R751 and placed under the control of the hepA promoter—functional only ...
example 3
Introduction of Recombinant DNA into F. heparinum
[0051] Conjugation:
[0052] Plasmids were introduced into the E. coli strain S17-1. S17-1 is a mobilizing strain which provides, in trans, the genes needed for conjugative DNA transfer.
[0053] Overnight cultures of both F. heparinum and E. coli S 17-1 containing the plasmid DNA were subcultured to an OD600 of 0.1 and grown to a mid-log phase of OD600 of 0.5. F. heparinum was grown in MG media supplemented with 1% heparin, 0.02% methionine / histidine, and 2 mM MgSO4 at 23° C. E. coli was grown in Lauria-broth supplemented with 50 μg / mL ampicillin and 10 μg / mL trimethoprim at 37° C.
[0054] 1.75 mL of F. heparinum and 100 μL of E. coli were combined in a 2 mL eppendorf tube and spun at 2500 g for 3 minutes. The cell pellet was resuspended in 100 μL MG and spread over a 0.45 mm membrane (Millipore, catalog number HAWG 025 00) which was placed on an agar petri dish (preheated for 30 minutes at 30° C.) containing LB / MG medium supplemented wi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| voltage | aaaaa | aaaaa |
| resistance | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com