Production of complex carbohydrates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Selection of a Hib Production Cell
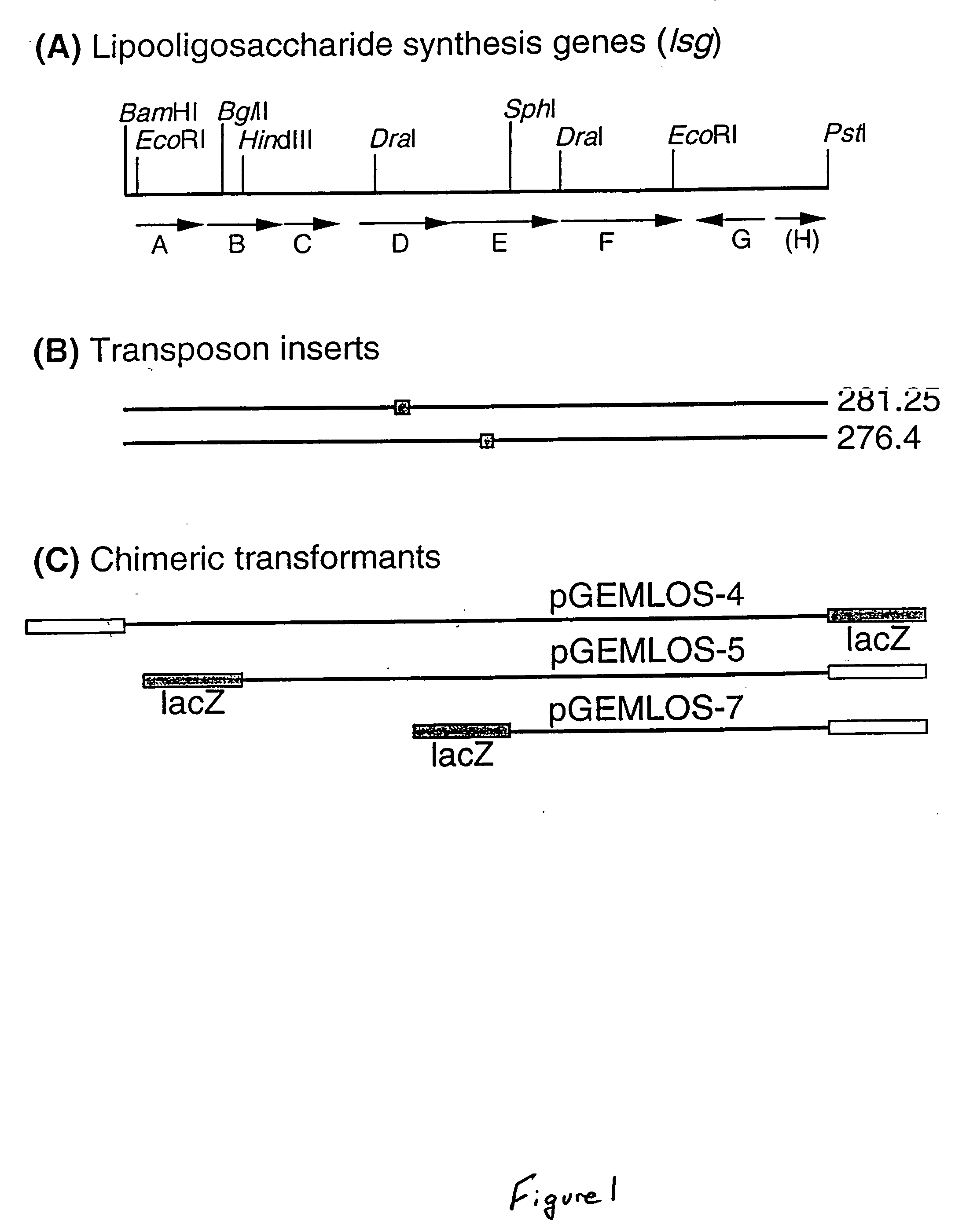
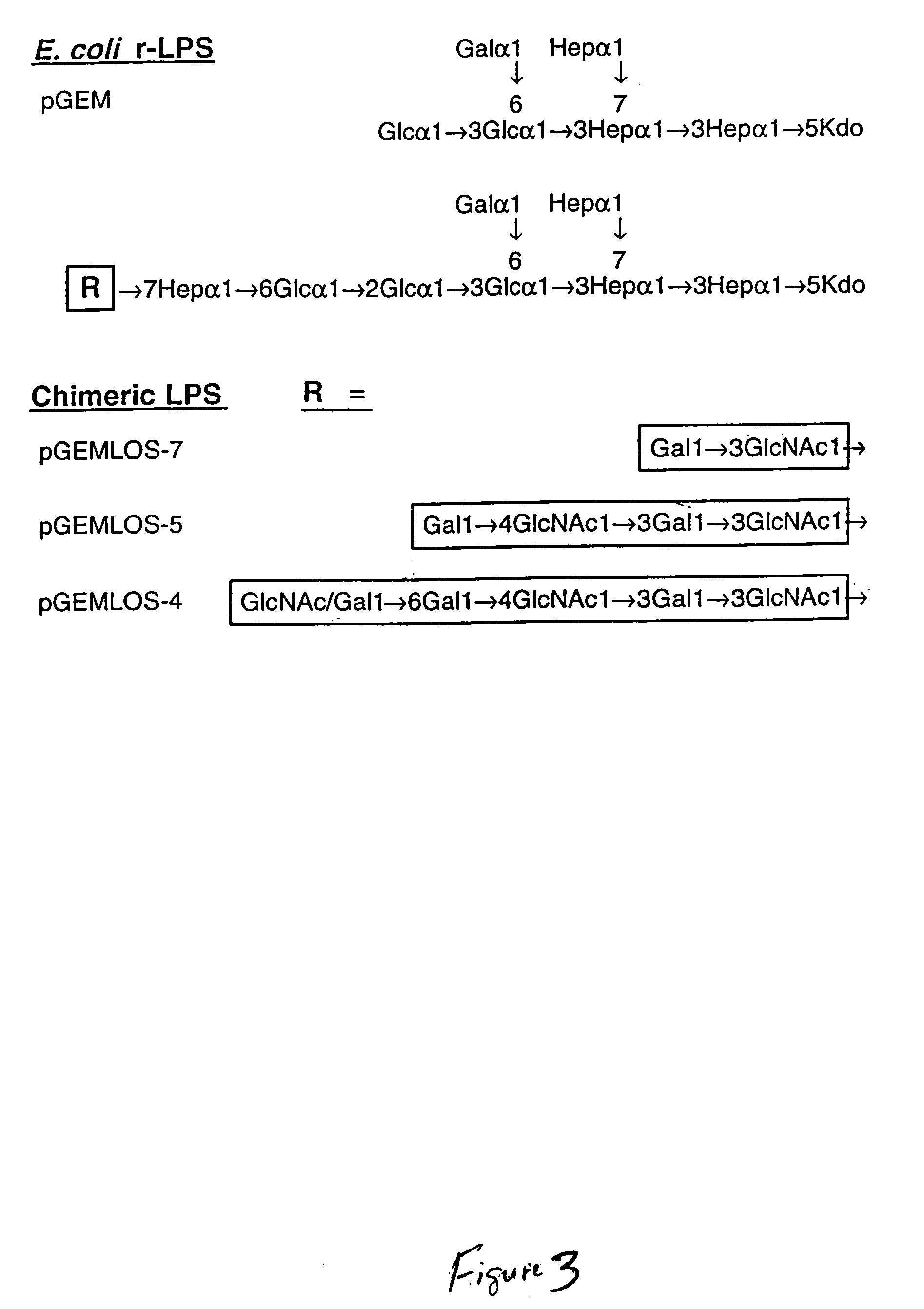
[0030] Capsular strains of Haemophilus influenzae type b (Hib) are responsible for various invasive and bacteraemic infections in humans, including meningitis and pneumonia. The surface lipooligosaccharides (LOS) of Hib are known to be important factors in microbial virulence and pathogenesis. Structural studies of Hib LOS from wild-type and mutant strains have shown that the LOS contains a conserved heptose trisaccharide core which can be extended with additional sugars on each heptose. Recently, a revised structure of the E. coli K-12 core region was reported which also contains a heptose trisaccharide inner core and a fourth heptose present on the terminus of the main oligosaccharide branch:
[0031] Previous work showed that the core region of E. coli transformed with synthetic enzyme genes could be elongated by the addition of saccharide monomers under the direction of H. influenzae genes to produce a modified E. coli LPS that was elongated by ...
example 3
Transformation of the Hib Production Cell
[0038] Restriction fragments of EMBLOS-I were used to make a series of plasmids which modified E. coli strain JM 109 to give clones which produced a proposed chimeric series of higher mass LPS species. The transformants termed pGEMLOS-4, pGEMLOS-5, and pGEMLOS-7 generated modified or chimeric LPS of 5.5, 5.1, and 4.5 kDa, respectively. All three apparently modified the 4.1 kDa LPS species from E. coli, although only the LPS from pGEMLOS-4 expressed the 6E4 epitope. The LPS from strain pGEMLOS-5 was found to react positively with MAb 3F 11, suggesting the presence of terminal N-acetyllactosamine. The epitope recognized by MAb 6E4 is also present in the LOS of H. influenzae nontypable strain 2019, as well as the LPS from Salmonella minnesota Re mutant. Binding of this monoclonal antibody to H. influenzae LOS can be inhibited by Kdo and the Kdo trisaccharide from the Re mutant of S. minnesota. Because the 6E4 epitope has been associated with th...
example 4
Isolation and Purification of Oligosaccharides
[0040] The LPS from PGEM (31 mg), pGEMLOS-4 (25 mg), pGEMLOS-5 (15 mg), and pGEMLOS-7 (4.4 mg) was hydrolyzed in 1% acetic acid (2 mg LPS / ml) for 2 hours at 100° C. The hydrolysates were centrifuged at 5000 g for 20 min at 4° C. and the supernatants removed. The pellets were washed with 2 ml of H2O and centrifuged again (5000 g, 20 min, 4° C.). The supernatants and washings were pooled and lyophilized to give the oligosaccharide fractions. As a standard, 10 mg of LPS from Salmonella typhimurium TV 119 Ra mutant (Sigma, St. Louis) was treated in the same fashion.
[0041] To prepare desalted oligosaccharide pools for ESI-MS analysis, small aliquots of the crude oligosaccharide fractions (<2 mg) were chromatographed on two Bio-Select SEC 125-5 HPLC columns (Bio-Rad, Richmond, Calif.) connected in series, using 0.05 M pyridinium acetate (pH 5.2) at a flow rate of 1 ml / min. A refractive index detector was used to monitor column effluent and c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Acceleration | aaaaa | aaaaa |
| Acceleration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com