Methods and Compositions Relating to Gene Silencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Preparation of an hsiRNA Mixture
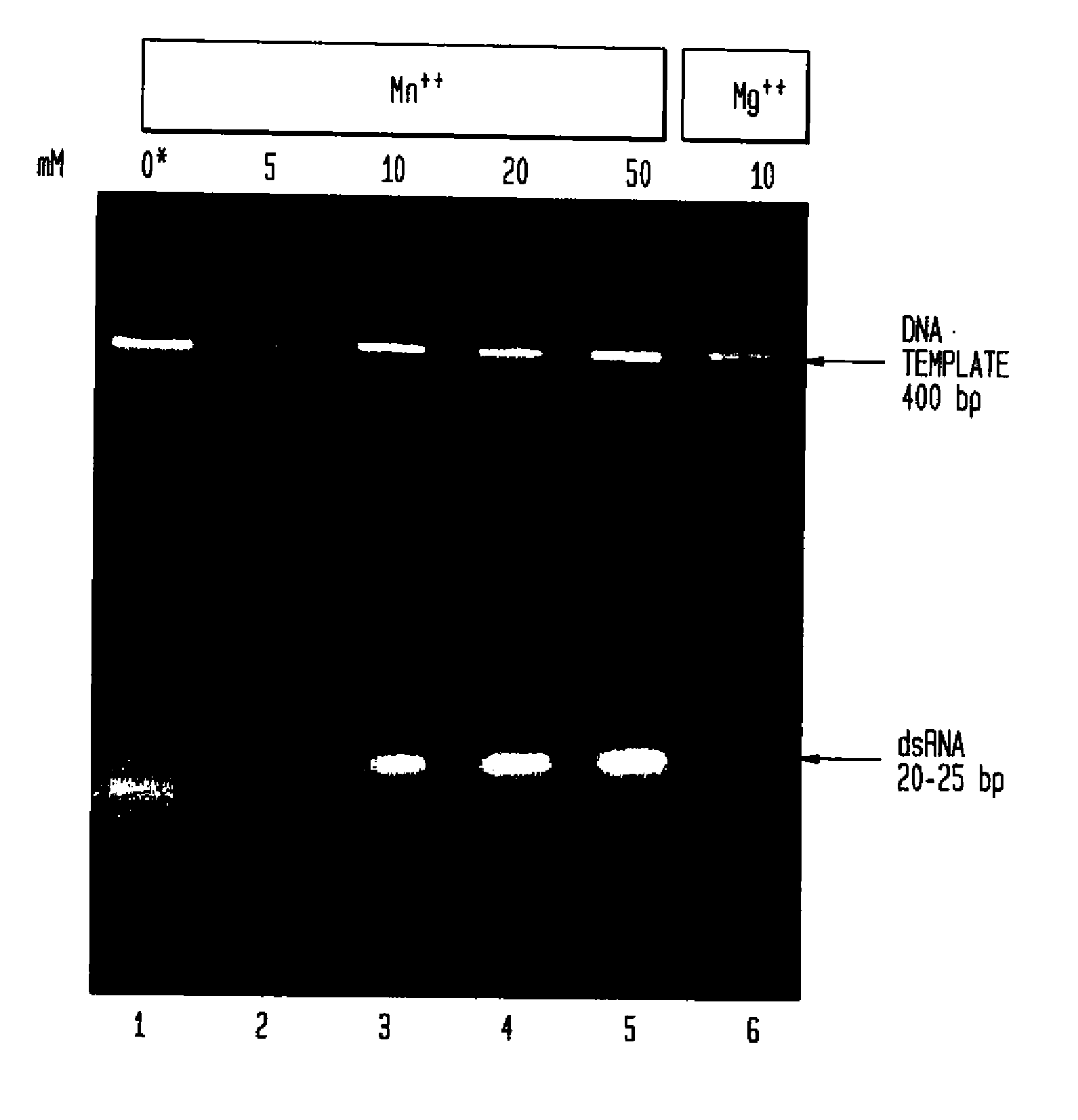
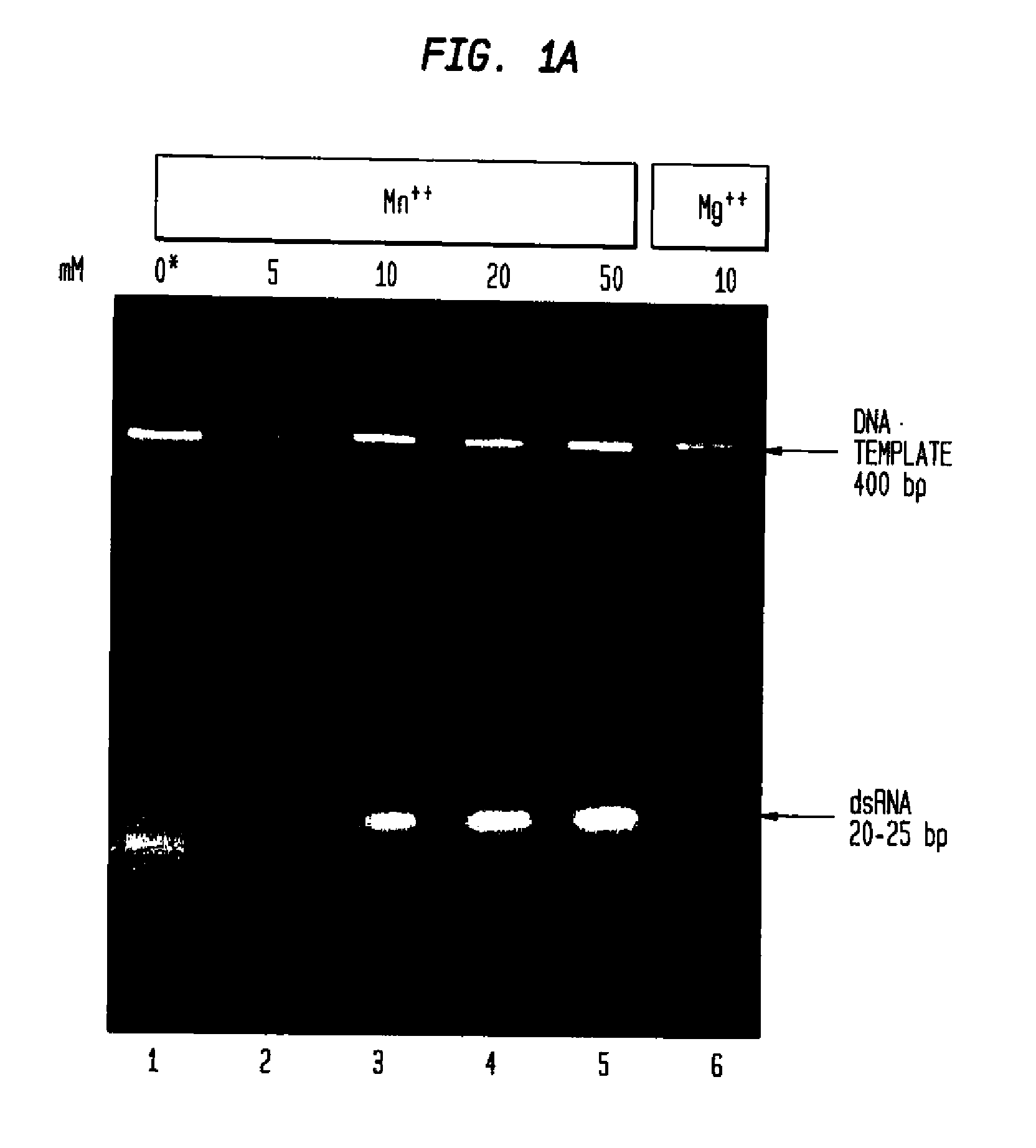
[0131]Determining the Effect of Manganese Ions on Cleavage of dsRNA by RNaseIII.
[0132]Full length double-stranded RNA (dsRNA) corresponding to the gene of interest, in this Example hu PKR, was generated using the HiScribe M RNAi Transcription Kit (New England Biolabs, Inc., Beverly, Mass.). Methods for creating double-stranded RNA are described in detail in Example VII.
[0133]A 0.4 kb double-stranded RNA molecule (0.25 μg) was digested with 30 units E. coli RNaseIII (0.9 μg) (New England Biolabs, Inc., Beverly, Mass.) in 20 μl of buffer consisting of 100 mM NaCl, 50 mM Tris-HCl, 5, 10, 20 or 50 mM MnCl2 or 10 mM MgCl2 (control), 1 mM dithiothreitol (pH 7.5@25° C.) at 37° C. Samples containing 50-100 mM MnCl2 are also tested to provide complete digestion of the long double-stranded RNA.
[0134]Digestion products of RNaseIII in the presence of various concentrations of manganese ion were enriched in the size range of 18-25 bp. The mixture of fragments obta...
example ii
Preparation of hsiRNA Using Various Divalent Metal Ions
[0144]To determine the effect of various divalent cations on the cleavage products of RNaseIII, the following experiment was undertaken: 1 μg of a large double-stranded RNA molecule (800 bp) was digested with each of two concentrations of E. coli RNaseIII (0.04 μg / μl or 0.02 μg / μl) at pH 7.5 (25° C.) in 50 μL of buffer containing 100 mM NaCl, 50 mM Tris-HCl, 1 mM dithiothreitol and either 10 mM MgCl2 at 37° C. (lanes 1 and 2), 10 mM MnCl2 (lanes 3 and 4) 10 mM CoCl2 (lanes 5 and 6) or 10 mM NiSO4 (lanes 7 and 8) for 30 minutes. The results are shown in FIG. 2. A double-stranded RNA product having an approximate size of 22 bp (within a range of 20 bp-40 bp) was produced by complete digestion of the large double-stranded RNA in the presence of 0.04 μg / μl RNaseIII and 10 mM manganese ions. Digestion with 0.04 μg / μl RNaseIII in the presence of 10 mM Mg2+ buffer absent manganese ions produced fragments which were smaller than the des...
example iii
Short Double-Stranded RNA Cleavage Products of RNaseIII Digestion Contain Sequences Representing the Entire Parent Sequence
[0145]The DNA template for transcription of p53 (1.1 kb fragment encoding amino acids 100-393) was digested with the restriction enzyme AciI and the resulting fragments separated on an agarose gel. The gel was ethidium-stained, photographed and subsequently transferred to a nylon membrane (Hybond® N+, Amersham, Piscataway N.J.).
[0146]Double-stranded RNA synthesized by in vitro transcription of the 1.1 kb fragment was digested with RNaseIII at a final concentration of 0.04 μg / μl in the presence of 10 mM Mn++ at pH 7.5 and 25° C. for 30 minutes as described in Example II and the products were separated on a 20% native polyacrylamide gel.
[0147]The products of the digestion (the hsiRNA mixture) were visualized by ethidium bromide staining and the fraction corresponding to about 21 bp was excised in a small gel slice and purified by electro-elution for 20 min in a sm...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com