Novel Method for Cloning Variable Domain Sequences of Immunological Gene Repertoire
a gene repertoire and variable domain technology, applied in the field of new cloning methods of immunological gene repertoire, can solve the problem of limited multiplexing ability of pcr-based amplification, and achieve the effect of increasing the size and diversity of the protein library
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Gene-Specific Oliggnucleotide and RNA Promoter-Linked Primer Selection
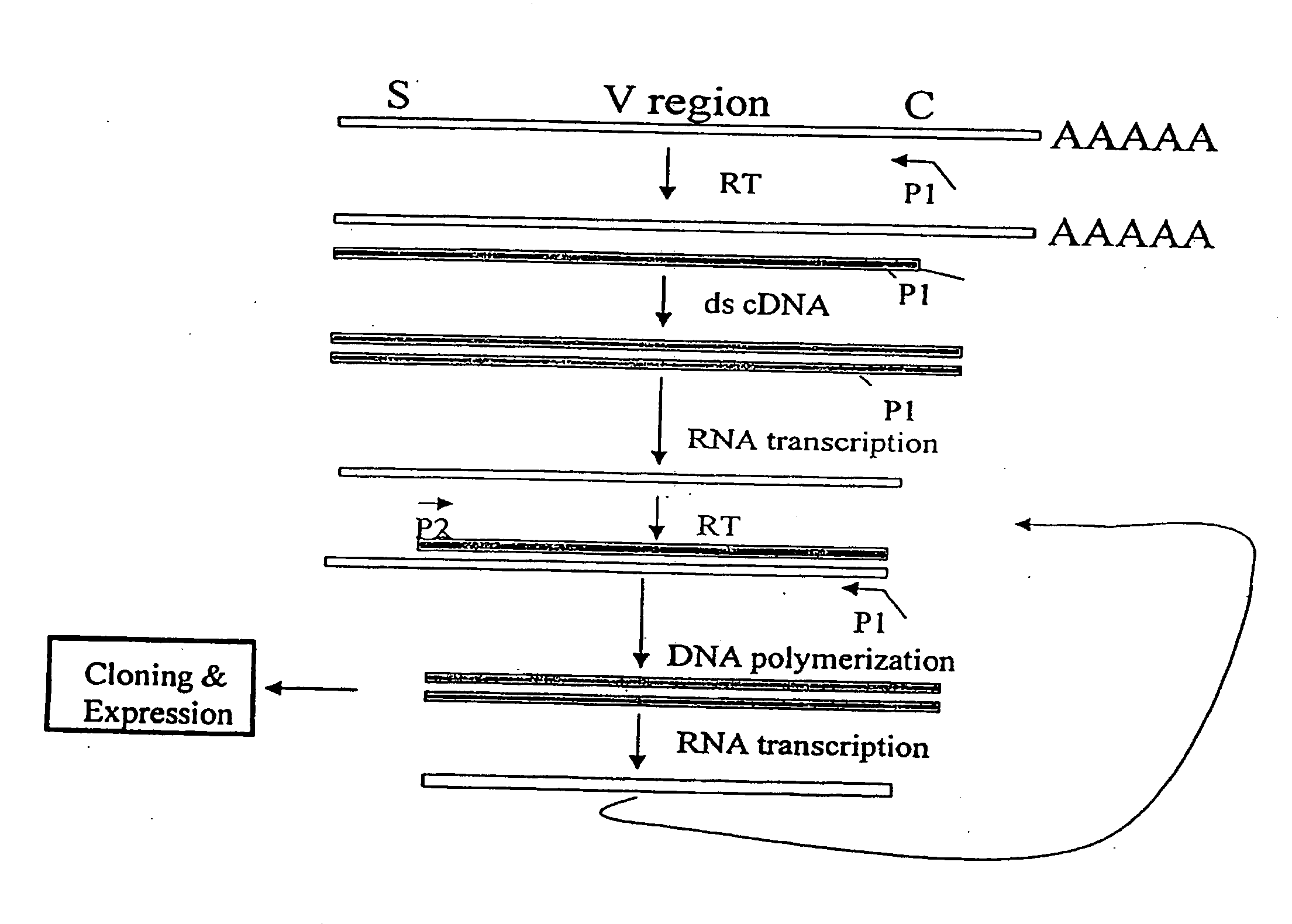
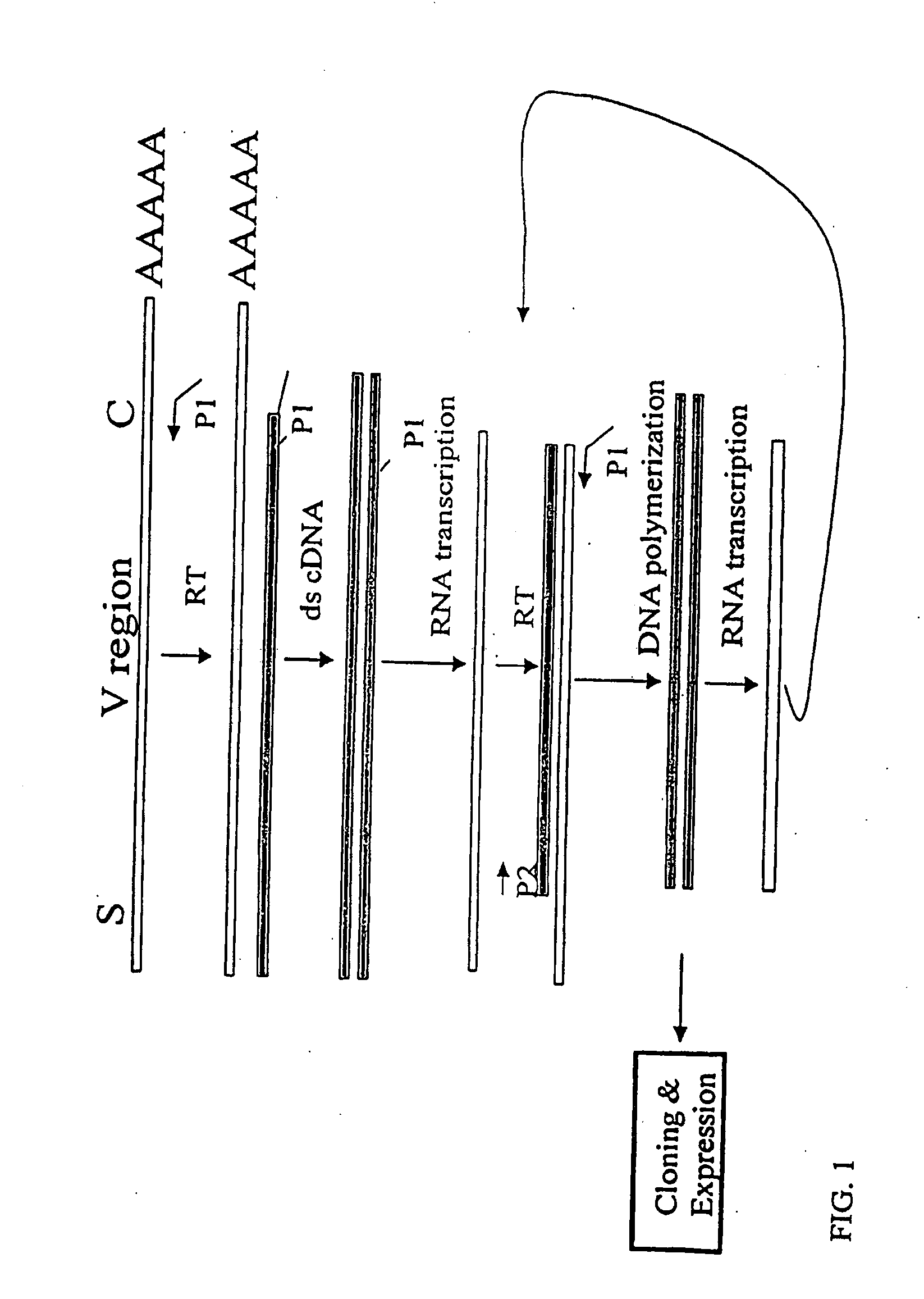
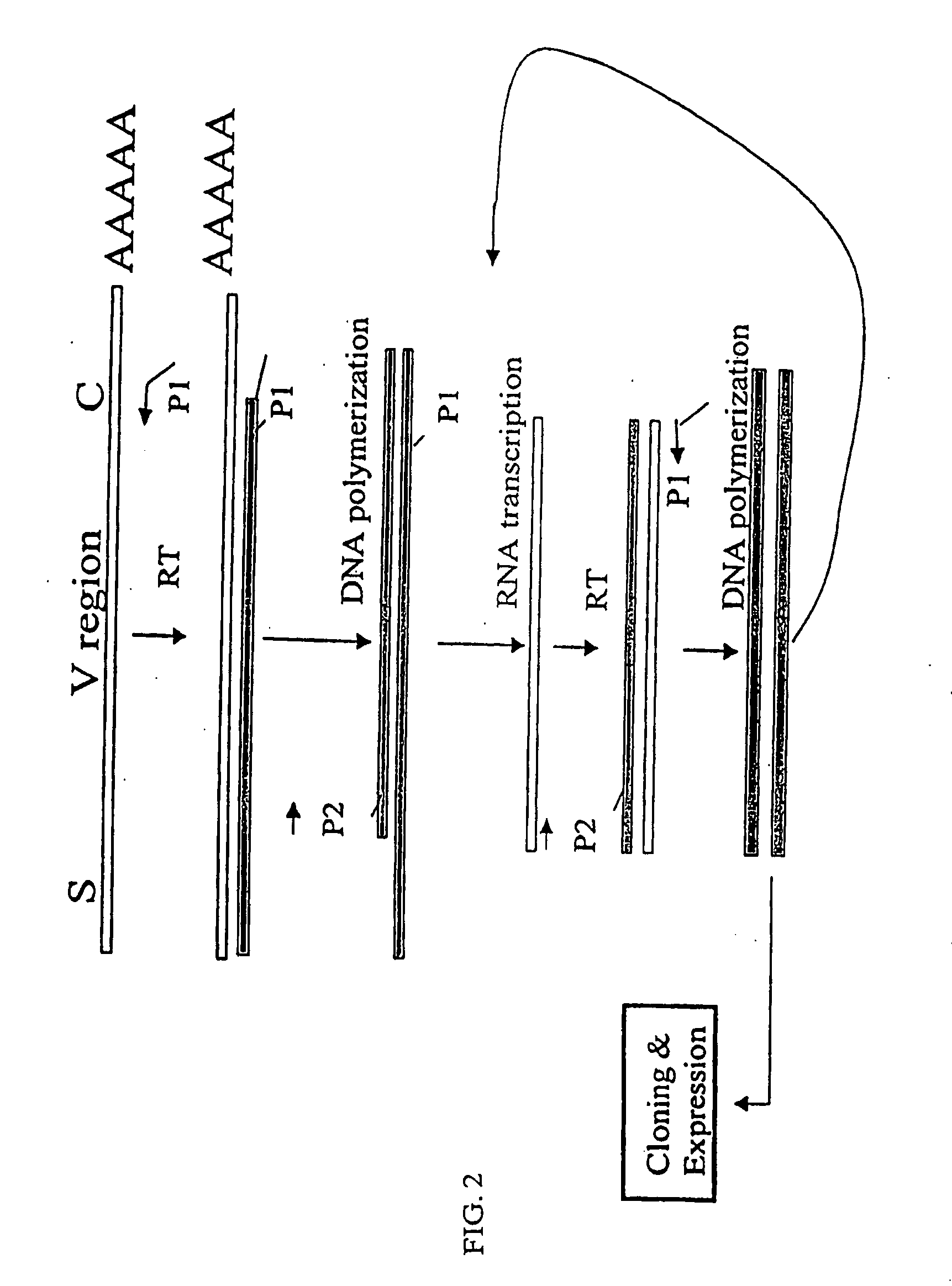
[0153]The nucleotide sequences coding for the human immunoglobulin complimentary determining region (CDR) are highly variable (Marks, J. D. et al., J. Mol. Biol., 222:581-597 (1991); Haidaris, C. G. et al., 257:185-202 (2001); Welschof, M. et al., J. Immunol. Methods, 179:203-214 (1995); Marks, J. D. et al., Eur. J. Immunol., 21:985-991 (1991); and Haard, H. J. D. et al., J. Biol. Chem., 274:18218-18230 (1999)). However, there are several regions of conserved sequences that flank the human VH domains, containing substantially conserved nucleotide sequences, i.e., sequences that will hybridize to the same primer sequence in a number of different genes. Therefore, gene-specific oligonucleotide primers can be selected for both gene-specific primers and the promoter-linked primers and synthesized to hybridize to the conserved sequences for reverse transcription, double stranding and RNA transcription reactions as desc...
example 2
Preparation of Source mRNAs Containing the VH and VL Gene Repertoire
[0160]Total cellular RNA was prepared from the blood cells collected from a pool of patients using the RNA preparation methods well known in the art as described by Chomczynski et al., Anal Biochem., 162:156-159 (1987) and the RNA isolation kit produced by QIAGEN GmbH (Hilden, Germany).
[0161]Messenger RNA (mRNA) enriched for sequences containing long poly A tracts was prepared from the total cellular RNA using methods described in Maniatis et al., eds., “Molecular Cloning: A Laboratory Press Manual”, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1982). Briefly, the total RNA isolated from the blood cells prepared as described above was resuspended in 1 ml of DEPC-H2O and maintained at 65° C. for 5 minutes. One ml of 2× high salt loading buffer consisting of 100 mM Tris-HCl, 1 M sodium chloride, 2.0 mM disodium ethylenediamine tetraacetic acid (EDTA) at pH 7.5, and 0.2% sodium dodecyl sulfate (SDS) w...
example 3
Transcriptional Amplification of the VH Gene Repertoire
[0163]Transcriptional amplification is performed using the scheme as depicted in FIG. 4. In detail, about 5-10 μg of poly(A)+ mRNAs in DEPC-treated water were first hybridized (annealed) with 1 μM antisense primer mixture comprising equal amounts of antisense EcoRI & stop codon-linked HJH primers, for example, aHJH-1, 5′-dTGG AAT GAA TTCGAT TGC TAGTCAGAC GGT GAC CAG GGT GCC-3′ (SEQ ID NO:_); aHJH-2, 5′-dTGG AAT GAA TTC GAT TGC TAG TCAGAC GGT GAC CAT TGT CCC-3′ (SEQ ID NO:_); aHJH-3, 5′-dTGG AAT GAA TTC GAT TGC TAG TCA GAC GGT GAC CAG GGT TCC-3′ (SEQ ID NO:_); and aHJH-4, 5′-dTGG AAT GAA TTC GAT TGC TAG TCAGAC GGT GAC CGT GGT CCC-3′ (SEQ ID NO:_) as listed in Table 1 (the EcoRI site and stop codons in three different frames are underlined), at 65° C. for 5 minutes and then cooled down to room temperature. The mixture was subsequently added to a reverse transcription (RT) reaction admixture (20 μl) on ice, comprising 2 μl of 10× b...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com