Improved negative-strand RNA viral vector
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
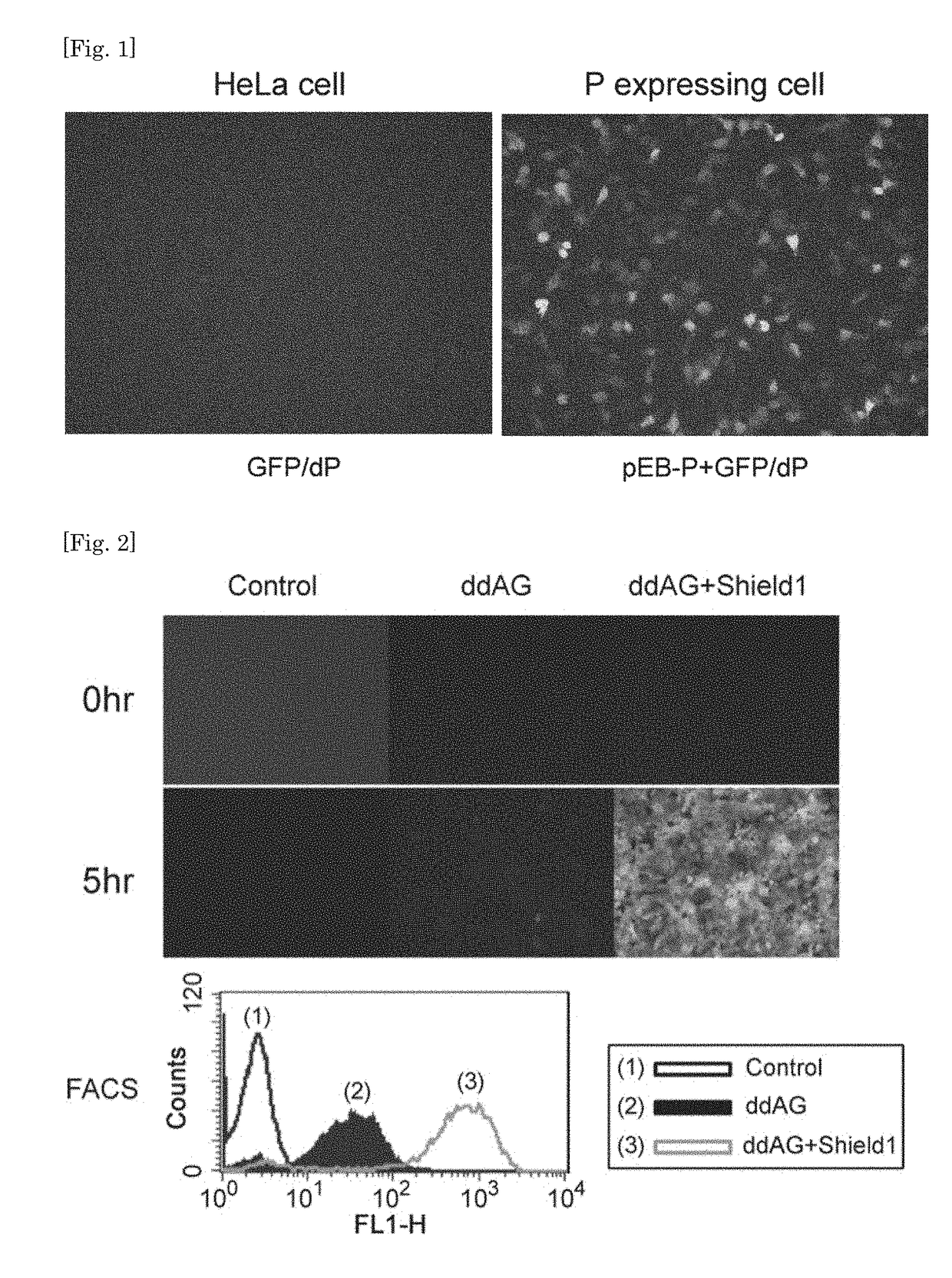
[0170]HeLa cells were seeded into 12-well plates at 1×105 cells / well. SeV(F)ddAG / TSΔF (ddAG) was infected at MOI=10 at 37° C. (day 0), 1 μM of Shield1 was added thereto on day 1, and after 5 hr. observation with a fluorescence microscope and analysis by FACS were carried out. FACScalibur (BD Biosciences) was used as FACS. As a result, green fluorescence was observed in ddAG infected cells added with Shield and strong fluorescence of ddAG+Shield1 was observed with FACS. It was difficult to determine fluorescence of ddAG not added with Shield1 under the microscope, but in FACS, fluorescence at the base level that is clearly discerned from control cells which were not infected with ddAG was observed (FIG. 2).
example 3
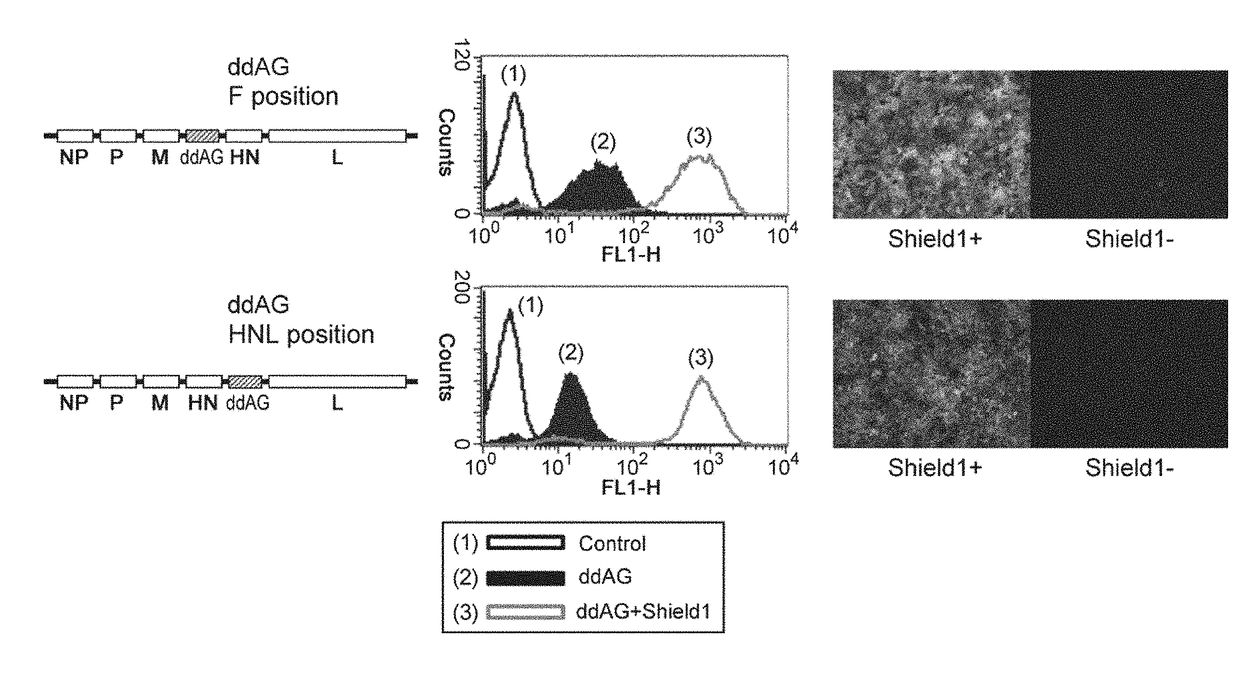
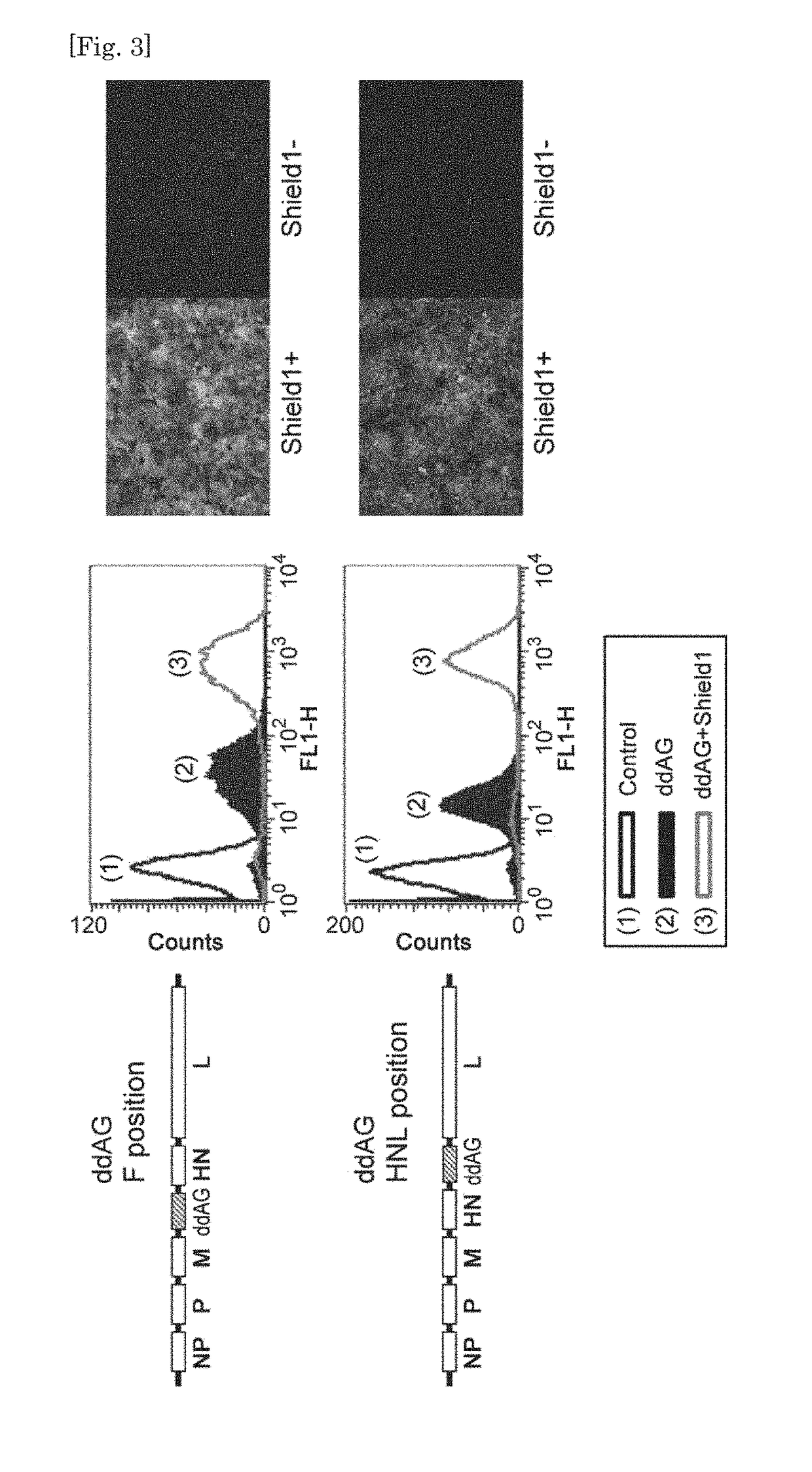
[0171]HeLa cells were infected with SeV(HNL)ddAG / TSΔF (ddAG) at MOI=10 at 37° C. (day 0), 1 μM of Shield1 was added thereto on day 1, and after 5 hr, observation with a fluorescence microscope and analysis by FACS were carried out. As a result, green fluorescence was observed in ddAG infected cells added with Shield1 and strong fluorescence of ddAG+Shield1 was observed with FACS. Fluorescence of ddAG not added with Shield was decreased by carrying the gene at HNL position as compared to SeV(F)ddAG / TSΔF on which the gene was carried at F position, but with FACS, fluorescence at the base level that is clearly discerned from control cells which were not infected with ddAG was observed (FIG. 3).
example 41
[0172]HeLa cells were infected with SeV(HNL)d1GFP / TSΔF (d1GFP), SeV(HNL)d2GFP / TSΔF (d2GFP), SeV(HNL)d4GFP / TSΔF (d4GFP), SeV(HNL)d1AG / TSΔF (d1AG), SeV(HNL)d2AG / TSΔF (d2AG), and SeV(HNL)d4AG / TSΔF (d4AG) at MOI=10 at 37° C. (day 0), and then observation with a fluorescence microscope was carried out on day 2. As a result, a decrease in fluorescence was observed by adding a PEST sequence to the C terminus side of GFP or AG Among d4GFP, d2GFP, and d1GFP, fluorescence of d4GFP was strong and fluorescence of d1GFP was weak (FIG. 4). It was possible to observe d2AG and d4AG under the fluorescence microscope, but it was difficult to determine d1AG under the fluorescence microscope. Thus, in the subsequent tests, d2 and d4 were used.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com