Methods and products for nucleic acid production and delivery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNA Synthesis
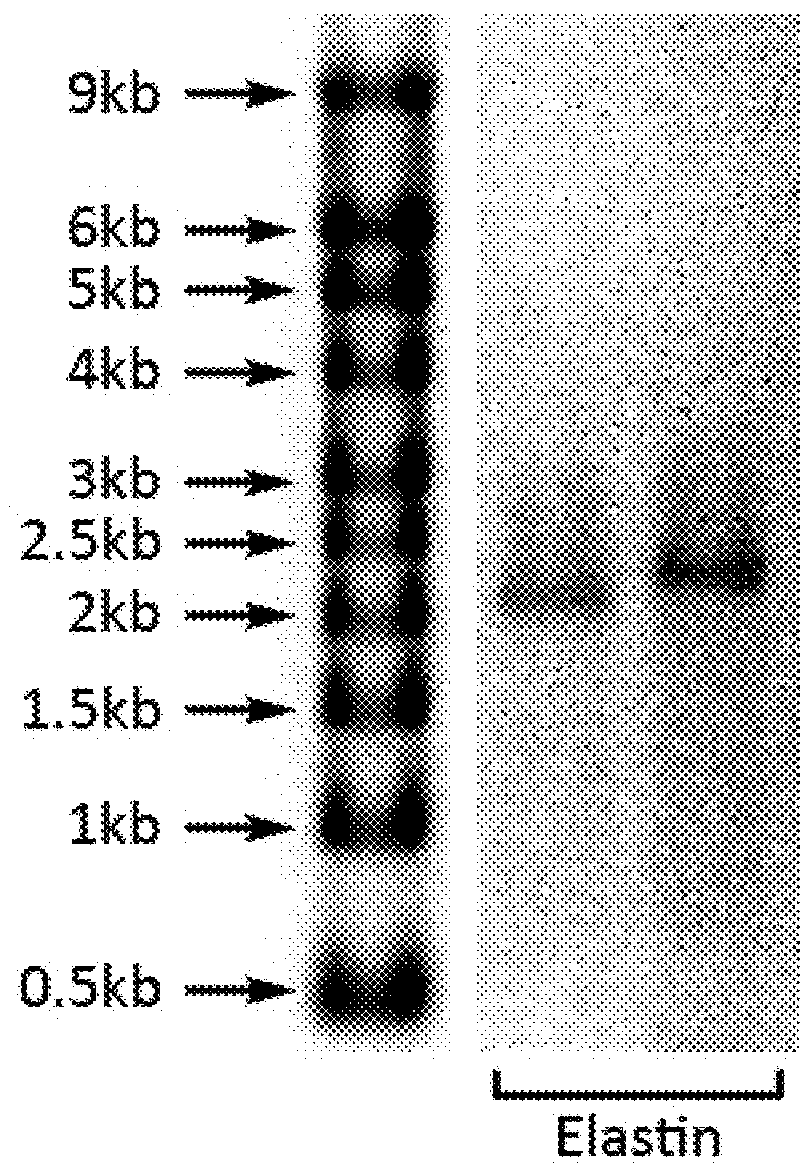
[0204]RNA encoding green fluorescent protein or the human proteins Elastin, Tyrosinase, Melanocortin 1 receptor, Hyaluronan synthase 1, Hyaluronan synthase 2, Hyaluronan synthase 3, Collagen type III al, Collagen type VII al, Interleukin 10, P-selectin glycoprotein ligand-1, Alpha-(1,3)-fucosyltransferase Oct4, Sox2, Klf4, c-Myc-2 (T58A), and Lin28 or TALENs targeting the human genes XPA, CCR5, TERT, MYC, and BIRC5, and comprising various combinations of canonical and non-canonical nucleotides, was synthesized from DNA templates using the T7 High Yield RNA Synthesis Kit and the Vaccinia Capping System kit with mRNA Cap 2′-O-Methyltransferase (all from New England Biolabs, Inc.), according to the manufacturer's instructions and the present inventors' previously disclosed inventions (U.S. application Ser. No. 13 / 465,490 (now U.S. Pat. No. 8,497,124), International Application No. PCT / US12 / 67966, U.S. application Ser. No. 13 / 931,251, and International Application No. PCT / U...
example 2
Transfection of Cells with Synthetic RNA
[0206]For transfection in 6-well plates, 2 μg RNA and 6 μL transfection reagent (Lipofectamine RNAiMAX, Life Technologies Corporation) were first diluted separately in complexation medium (Opti-MEM, Life Technologies Corporation or DMEM / F12+10 μg / mL insulin+5.5 μg / mL transferrin+6.7 ng / mL sodium selenite+2 μg / mL ethanolamine) to a total volume of 60 μL each. Diluted RNA and transfection reagent were then mixed and incubated for 15 min at room temperature, according to the transfection reagent-manufacturer's instructions. Complexes were then added to cells in culture. Between 12 μL and 240 μL of complexes were added to each well of a 6-well plate, which already contained 2 mL of transfection medium per well. Plates were shaken gently to distribute the complexes throughout the well. Cells were incubated with complexes for 4 hours to overnight, before replacing the medium with fresh transfection medium (2 mL / well). Volumes were scaled for transfe...
example 3
Toxicity of and Protein Translation from Synthetic RNA Containing Non-Canonical Nucleotides
[0207]Primary human fibroblasts were transfected according to Example 2, using RNA synthesized according to Example 1. Cells were fixed and stained 20-24 h after transfection using an antibody against Oct4. The relative toxicity of the RNA was determined by assessing cell density at the time of fixation.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com