Novel guide rna/cas endonuclease systems
a technology of endonuclease and guide, which is applied in the field of plant molecular biology, can solve the problems of low specificity, costly and time-consuming preparation, and achieve the effect of reducing the number of endonuclease residues, and improving the specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
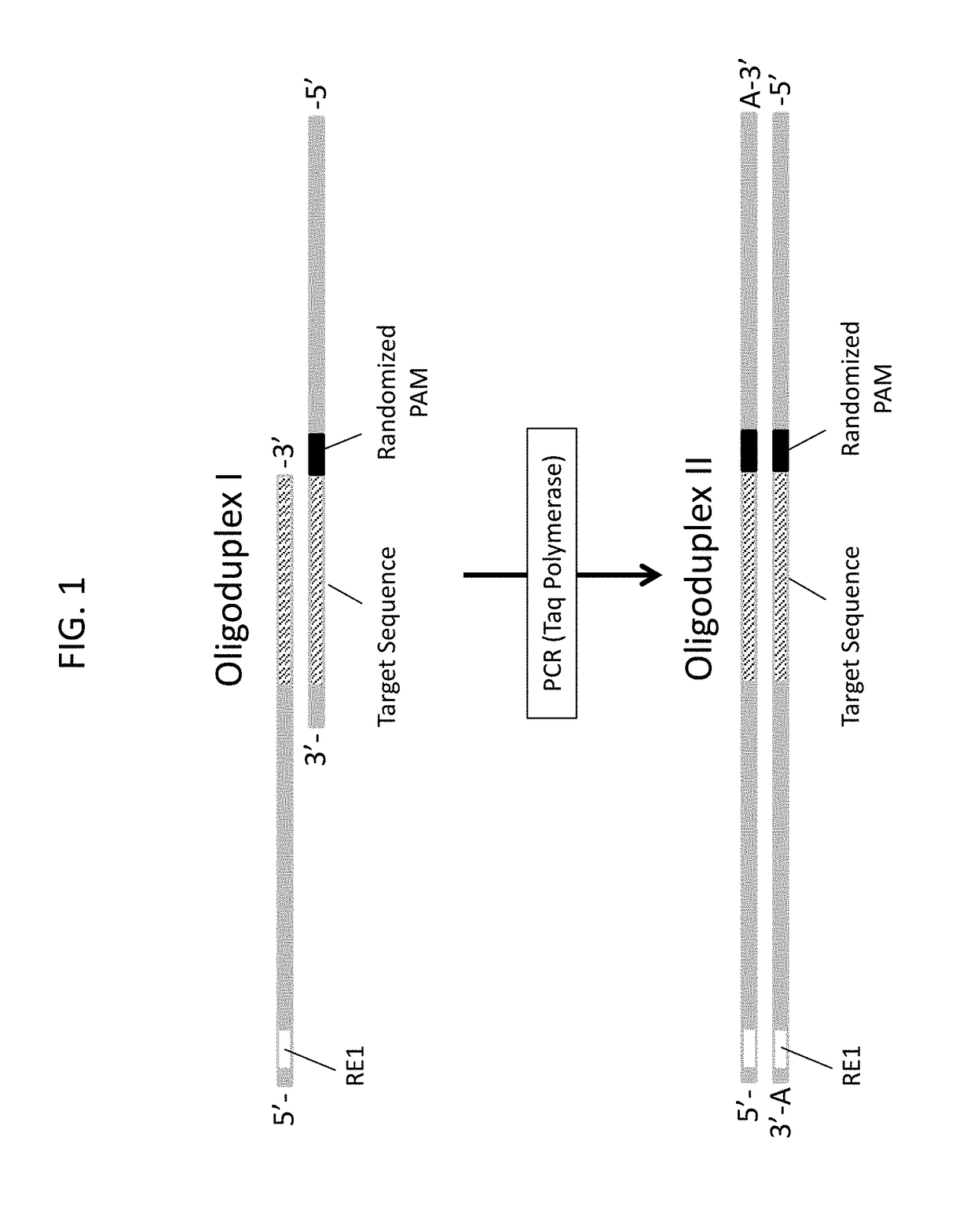
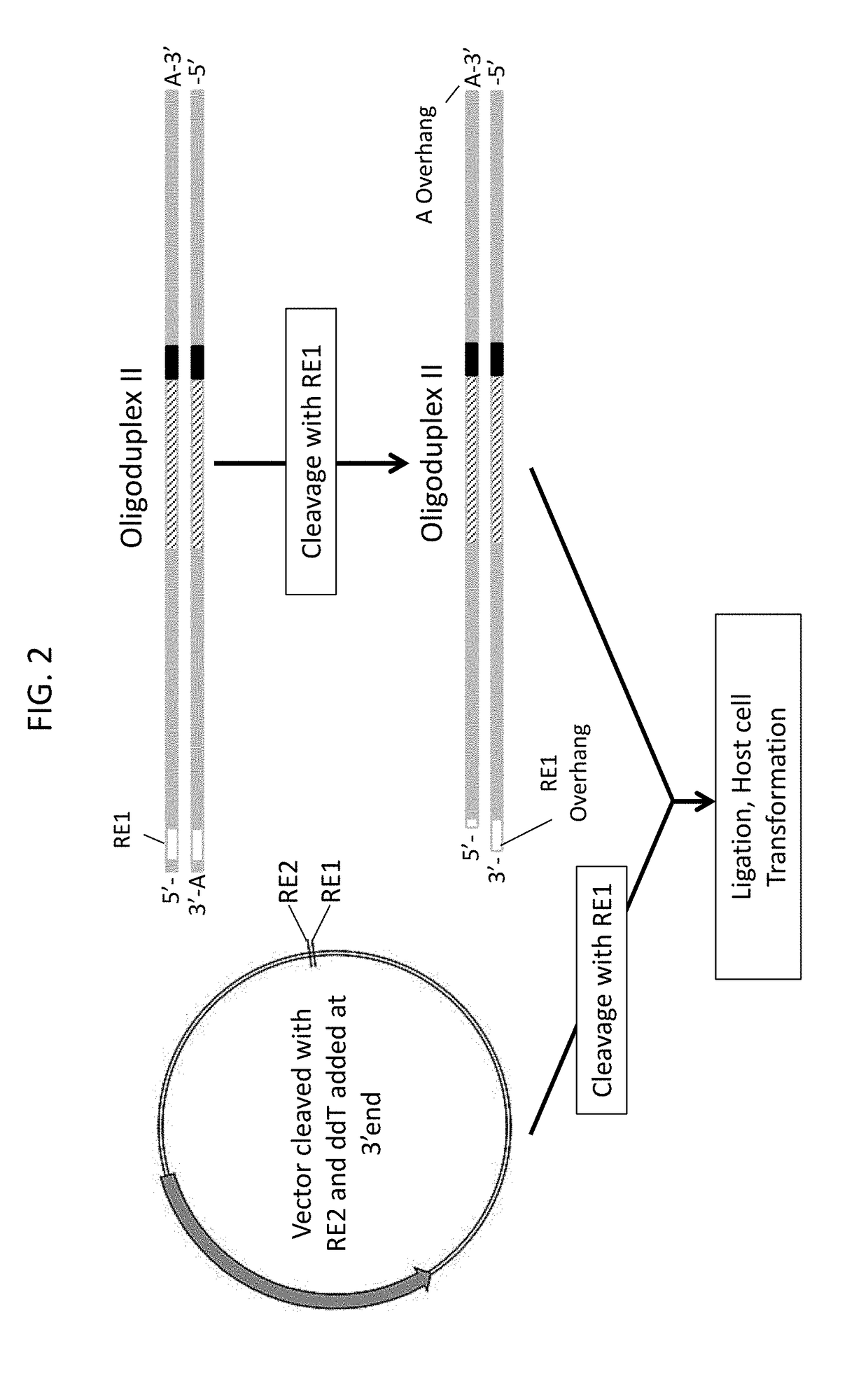
Design and Construction of 5N Randomized Protospacer-Adjacent-Motif (PAM) Library for Assaying Cas9 PAM Preferences
[0375]To characterize the Protospacer-Adjacent-Motif (PAM) specificity of Cas9 proteins from Type II CRISPR (clustered, regularly interspaced, short palindromic repeats)-Cas (CRISPR-associated) nucleic acid-based adaptive immune systems found in most archaea and some bacteria, a plasmid DNA library containing a section of 5 random base pairs immediately adjacent to a 20 base pair target sequence, T1 (CGCTAAAGAGGAAGAGGACA (SEQ ID NO: 1), was developed. Randomization of the PAM sequence was generated through the synthesis of a single oligonucleotide, GG-821N (TGACCATGATTACGAATTCNNNNNTGTCCTCTTCCTCTTTAGCGAGC (SEQ ID NO: 2), with hand-mixing used to create a random incorporation of nucleotides across the 5 random residues (represented as N in the sequence of GG-821N). To convert the single stranded template of GG-821N into a double-stranded DNA template for cloning into the ...
example 2
Protein Expression and Purification of Streptococcus pyogenes, Streptococcus Thermophilus CRISPR1 and Streptococcus thermophilus CRISPR3 Cas9 Proteins
[0377]To examine the PAM specificity of the Cas9 proteins from the Streptococcus pyogenes (Spy) (Jinek et al. (2012) Science 337:816-21), Streptococcus thermophilus CRISPR1 (Sth1) (Horvath et al. (2008) Journal of Bacteriology 190:1401-12) and Streptococcus thermophilus CRISPR3 (Sth3) (Horvath et al. (2008) Journal of Bacteriology 190:1401-12) Type II CRISPR-Cas systems, Spy, Sth1 and Sth3 Cas9 proteins were E. coli expressed and purified. Briefly, the cas9 genes of the CRISPR1-Cas and CRISPR3-Cas systems of Streptococcus thermophilus (Sth1 and Sth3) were amplified from a genomic DNA sample, while the cas9 gene of Streptococcus pyogenes (Spy) was amplified from a plasmid, pMJ806 (Addgene plasmid #39312)). DNA fragments encoding Sth1, Sth3 and Spy Cas9 were PCR amplified using Sth1-dir / Sth1-rev (ACGTCTCACATGACTAAGCCATACTCAATTGGAC (SEQ I...
example 3
Identification of PAM Preferences for Streptococcus pyogenes and Streptococcus thermophilus CRISPR3 Cas9 Proteins
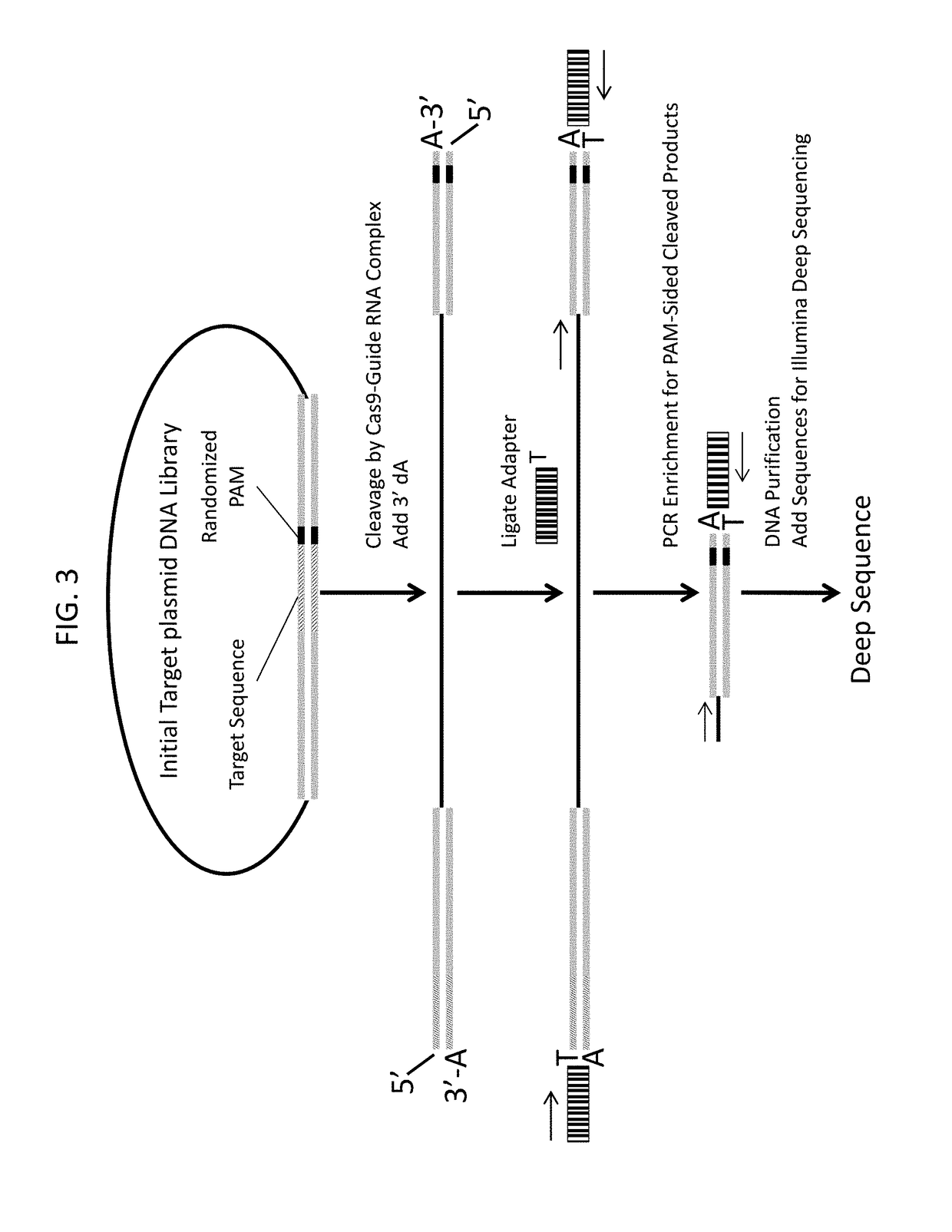
[0379]To empirically examine the PAM preferences for Streptococcus pyogenes (Spy) and Streptococcus thermophilus CRISPR3 (Sth3) Cas9 proteins, the randomized PAM library described in Example 1 was subject to digestion with purified Sth3 and Spy Cas9 proteins and guide RNA containing a variable targeting domain that hybridizes with, i.e., is complementary to, a sequence in the target DNA molecule (referred herein as target sequence), T1 (SEQ ID NO: 1). Sth3 and Spy Cas9-crRNA-tracrRNA complexes were assembled by mixing Cas9 protein with pre-annealed crRNA and tracrRNA duplex (Table 3) at 1:1 molar ratio followed by incubation in a complex assembly buffer (10 mM Tris-HCl pH 7.5 at 37° C., 100 mM NaCl, 1 mM EDTA, 1 mM DTT) at 37° C. for 1 h. 1 μg of plasmid DNA library with randomized 5 bp NNNNN PAM was cleaved with 50 nM and 100 nM of Cas9 complex in a reaction buffer (10 m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com