New CkNHX gene and shearing decorative gene CkNHXn, method for cultivating inverse-resistant plant
A plant and gene technology, applied in the field of the new CkNHX gene and its splicing modification gene CkNHXn, and the cultivation of stress-tolerant plants, can solve problems such as not yet retrieved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
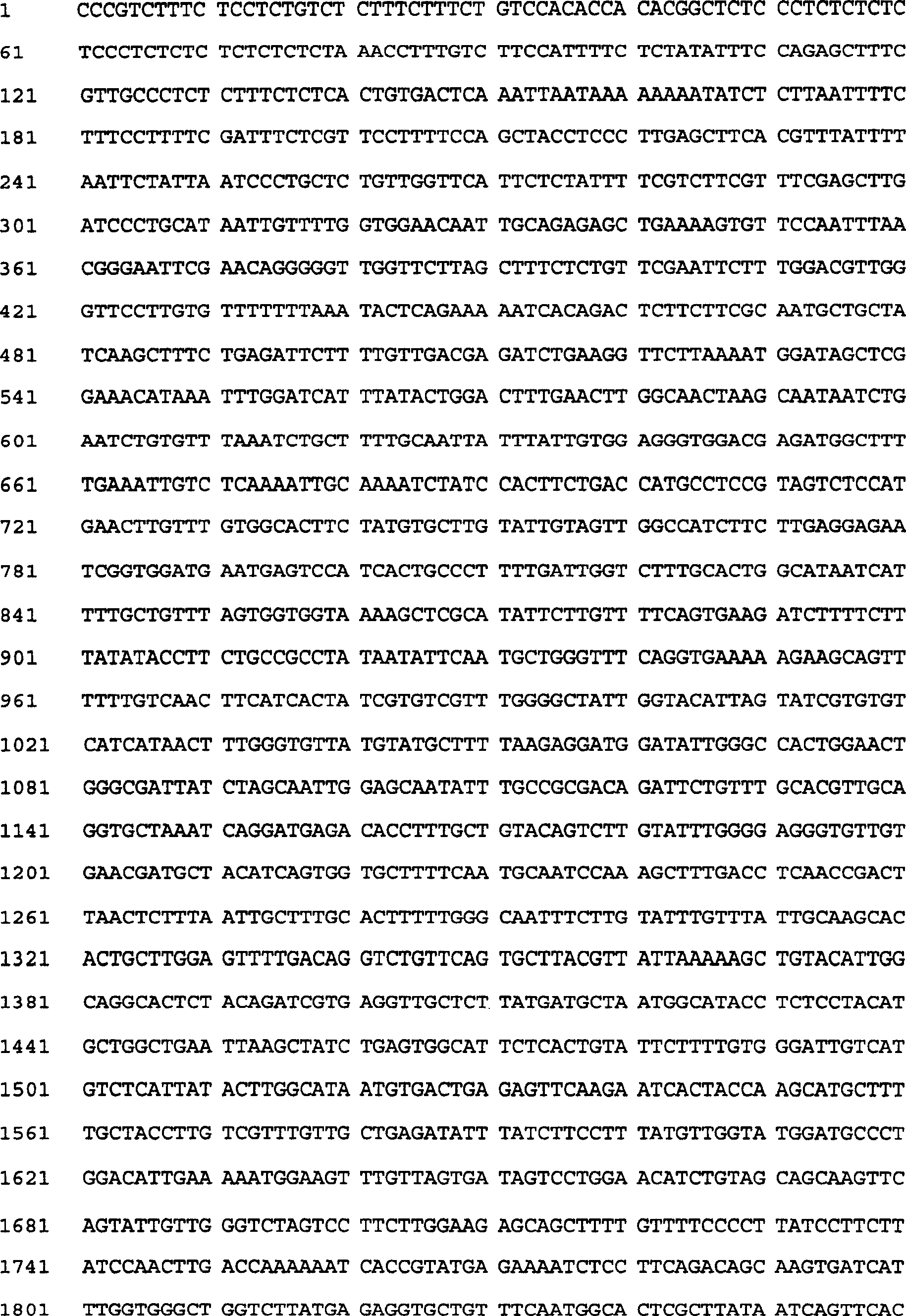
[0034] Embodiment 1, the cloning of Caragana CkNHX gene
[0035] Caragana seedlings grown for 2-3 months (purchased from germinated Caragana seeds in Xinjiang Turpan Desert Botanical Garden, Chinese Academy of Sciences) were used as materials, and induced overnight with 100uM ABA (Sigma Company). Take the leaves, wash them, put them into a DEPC-treated and sterilized homogenizer, add TRIzol (Invitrogen) to grind and homogenate, leave at room temperature for 5 minutes, add chloroform to extract twice, take the supernatant, and use iso Total RNA was precipitated with propanol, air-dried, and dissolved in an appropriate amount of DEPC water. cDNA was obtained by reverse transcription with polyT primers.
[0036] The primer sequence of polyT is as follows:
[0037] Oligo dT-Primer: 5'-CGAGCGGCCGCCCGGGCAGGTTTTTTTTTTTTTTTT-3' (SEQ ID NO: 5)
[0038] The RT reaction conditions are as follows:
[0039]
[0040] Incubate at 65°C for 10 minutes, immediately quench on ice, then ad...
Embodiment 2
[0086] Embodiment 2, the construction of CkNHXn gene
[0087] References reported (Yamaguchi T, Apse M P, Shi H Z, and Blumwald E(2003).Topological analysis of a plant vacuolar Na+ / H+ antiporter reveals luminal C terminus that regulates antiporter cation selectivity.PNAS.100(21):12510-12515 ), using the following primers to add a stop codon after the 435th amino acid to form a C-terminal truncated CkNHXn (that is, to remove the peptide after the 435th position) (see image 3 ). The primers used are:
[0088] The forward primer is (including Sac I site)
[0089] 5'-CGCGCC GAGCTC ATGGCTTTTGAAATTGTCT-3' (SEQ ID NO: 18)
[0090] The reverse primer is (including Xba I site)
[0091] 5'-CCGCGC TCTAGA CTAAGTCATCAAAACCAAACACC-3' (SEQ ID NO: 19)
[0092] High-fidelity Pyrobest Taq was used to perform PCR amplification using the above-mentioned reverse transcription product as a template, and the method was the same as before. The CkNHXn gene (SEQ ID NO: 3) with a fragment size ...
Embodiment 3
[0093] Example 3, used for yeast functional complementation experiments
[0094] 1. Construction of yeast expression vector
[0095] According to conventional molecular biology methods (F. Osper et al., translated by Yan Ziying and Wang Hailin, finely edited Molecular Biology Experiment Guide. Science Press, 2001), respectively introduce The SacI and XbaI sites were double-digested with SacI and XbaI, and then ligated into pYES2.0 (Invortrogene) double-digested with the same enzyme to construct two yeast expression vectors, pYES-NHX and pYES-NHXn. Its graph is constructed as Figure 8 , 9 As shown, after enzyme digestion, PCR identification and sequencing to ensure that the inserted fragment is correct, it was transferred into the salt-sensitive yeast mutant ANT3 (see references for details: Quintero FJ, Blattb MR, and Pardo JM (2000). Functional conservation between yeast and plant endosomal Na + / H + antiporters. FEBS Lett 471, 224-228). For specific operation, refer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com