Oligonucleotide for detecting cytochrome P450 enzyme series mutation site and gene chip
A technology of oligonucleotide probes and detection probes, which is applied in the field of nucleic acid detection of DNA genotypes, can solve problems such as the inability to determine mutation sites and mutation forms, high technical requirements for operators, and difficulty in popularization, achieving remarkable practicality value, the effect of preventing side effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1. Screening and optimization of oligonucleotide probes for detecting eight SNP sites of CYP450
[0034] 1. Preparation of oligonucleotide gene chips and design and screening optimization of oligonucleotide probes
[0035] The probes are respectively designed on the DNA sequences of each subtype of the human CYP450 enzyme system, and have high sensitivity and specificity. The oligonucleotide probe sequence is consistent with the base sequence of the DNA positive strand in the specific region of the above gene. When the probe is synthesized, its 3' terminal amino group is modified, which is used for the probe to react with the active aldehyde group on the surface of the slide and fix it to the surface of the carrier. After the candidate oligonucleotide probes are purified and quantified, they are spotted on the aldehyde-activated glass slides with a gene chip spotter to make a probe microarray.
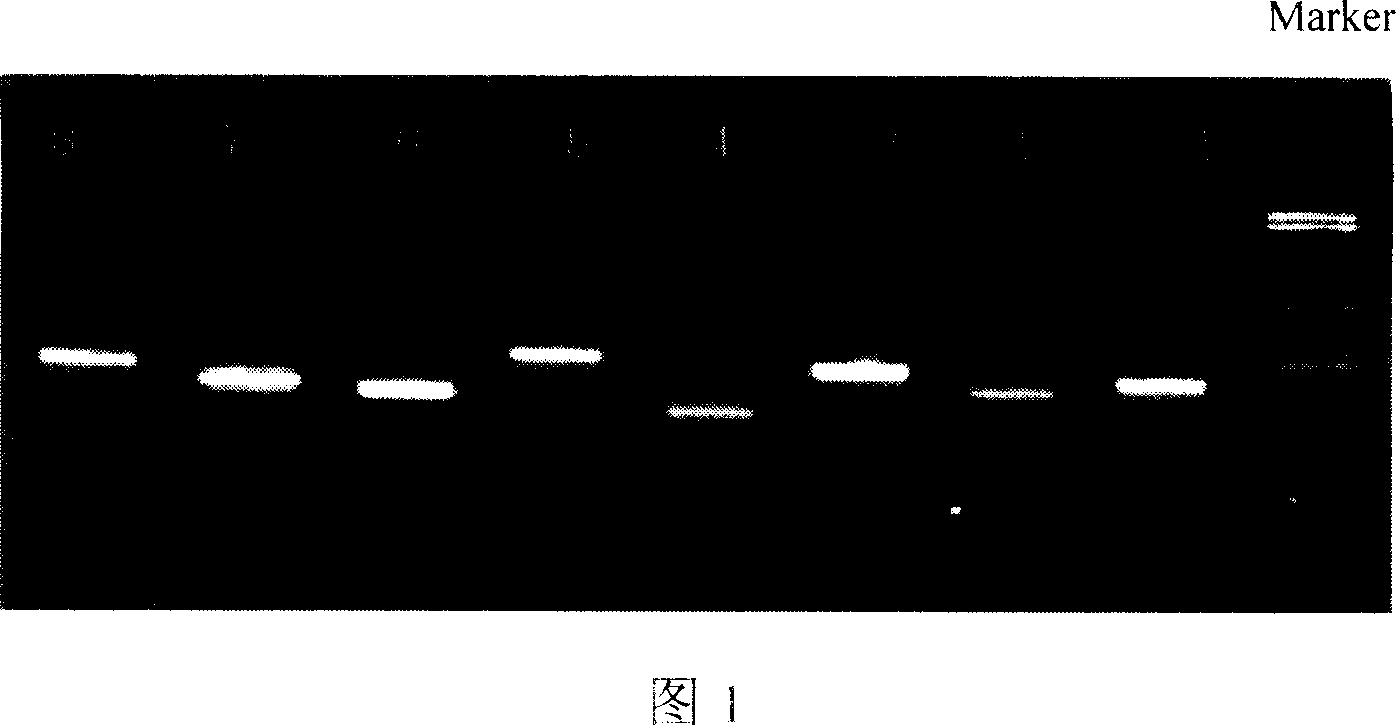
[0036] The PCR product using genomic DNA as a template was purifie...
Embodiment 2
[0041] Example 2. Specificity and Sensitivity Evaluation of Oligonucleotide Probes
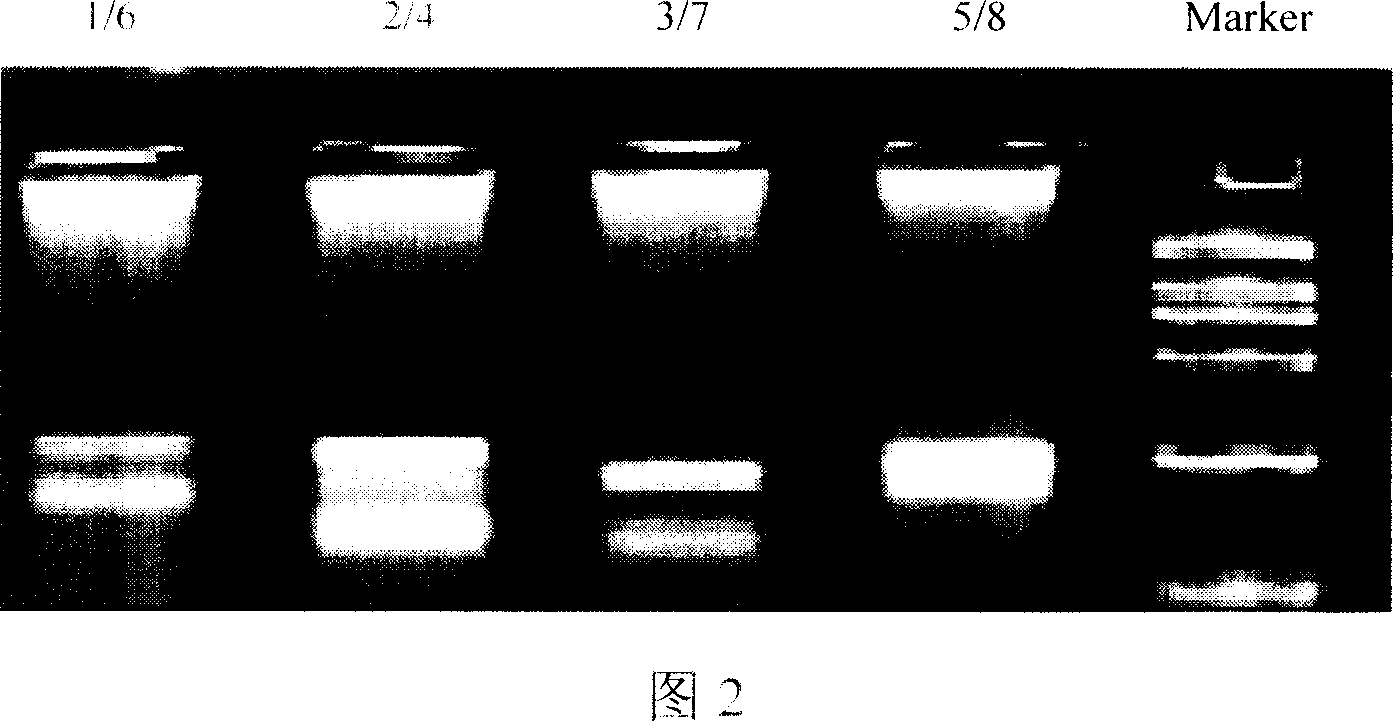
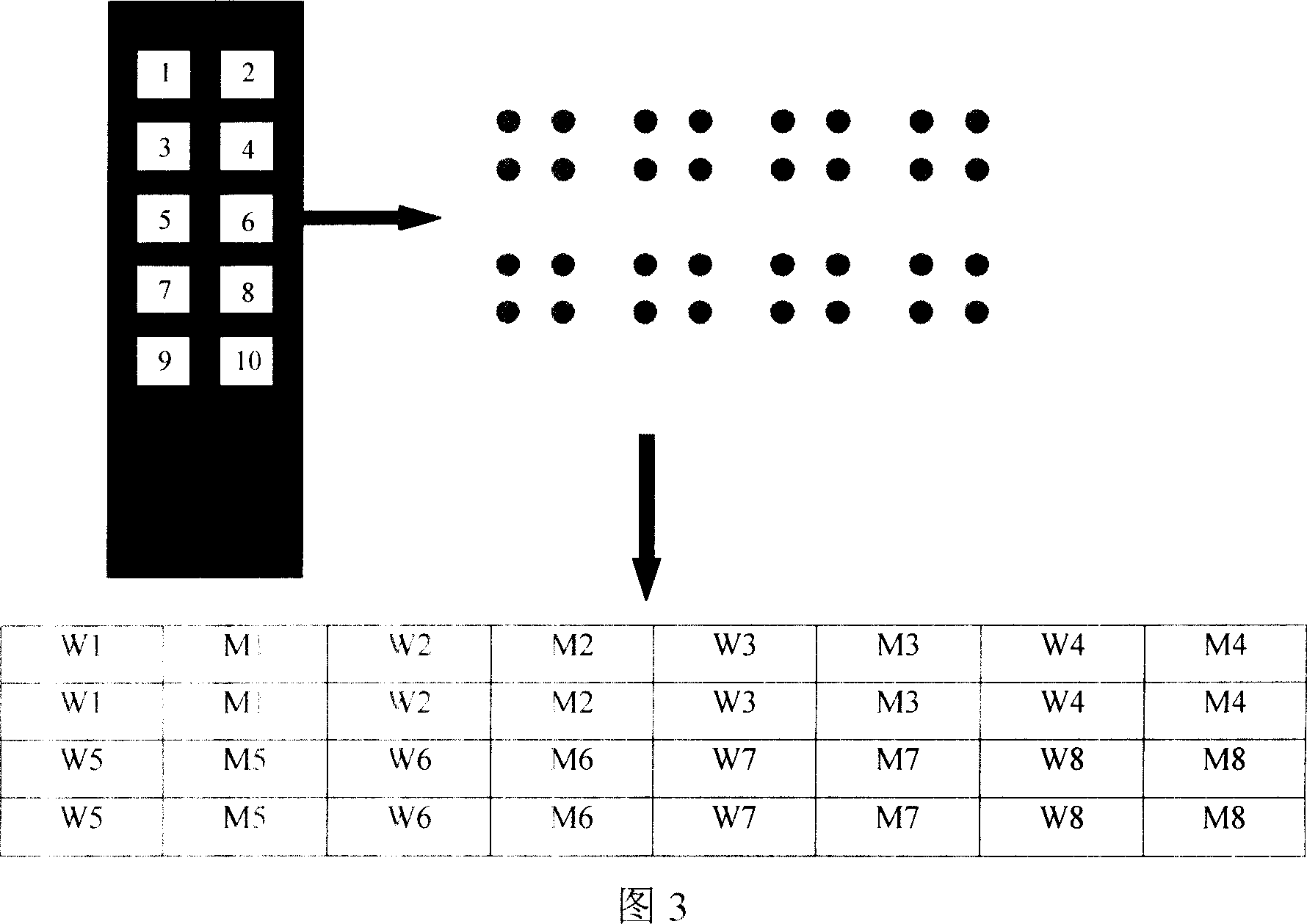
[0042] 1. Gene chip preparation Dilute the oligonucleotide probes with spotting solution (6×SSC, 0.1% SDS) to a final concentration of 50 μmol / L, and transfer 5ul to a 384-well plate. The probes were spotted on the aldehyde-based chip with a Cartesian chip preparation instrument (see Figure 3 for the arrangement of the probes, and see Figure 5 for the hybridization results of the chips with fluorescently labeled PCR products of CYP450 enzyme genomic DNA).
[0043] Specific detection of hotspot mutation site CYP450 oligonucleotide chip
[0044] Under the optimized conditions of two sets of double PCR amplification and hybridization reaction, 36 clinical DNA samples were detected, and four sets of double PCR amplification were carried out with the DNA samples as templates, and the products were hybridized with the chip. The probes in which signals appear correspond to the fragments of their hyb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com