Vectors comprising novel regulatory elements
一种载体、元件的技术,应用在重组DNA领域,能够解决5-甲基胞嘧啶不稳定等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
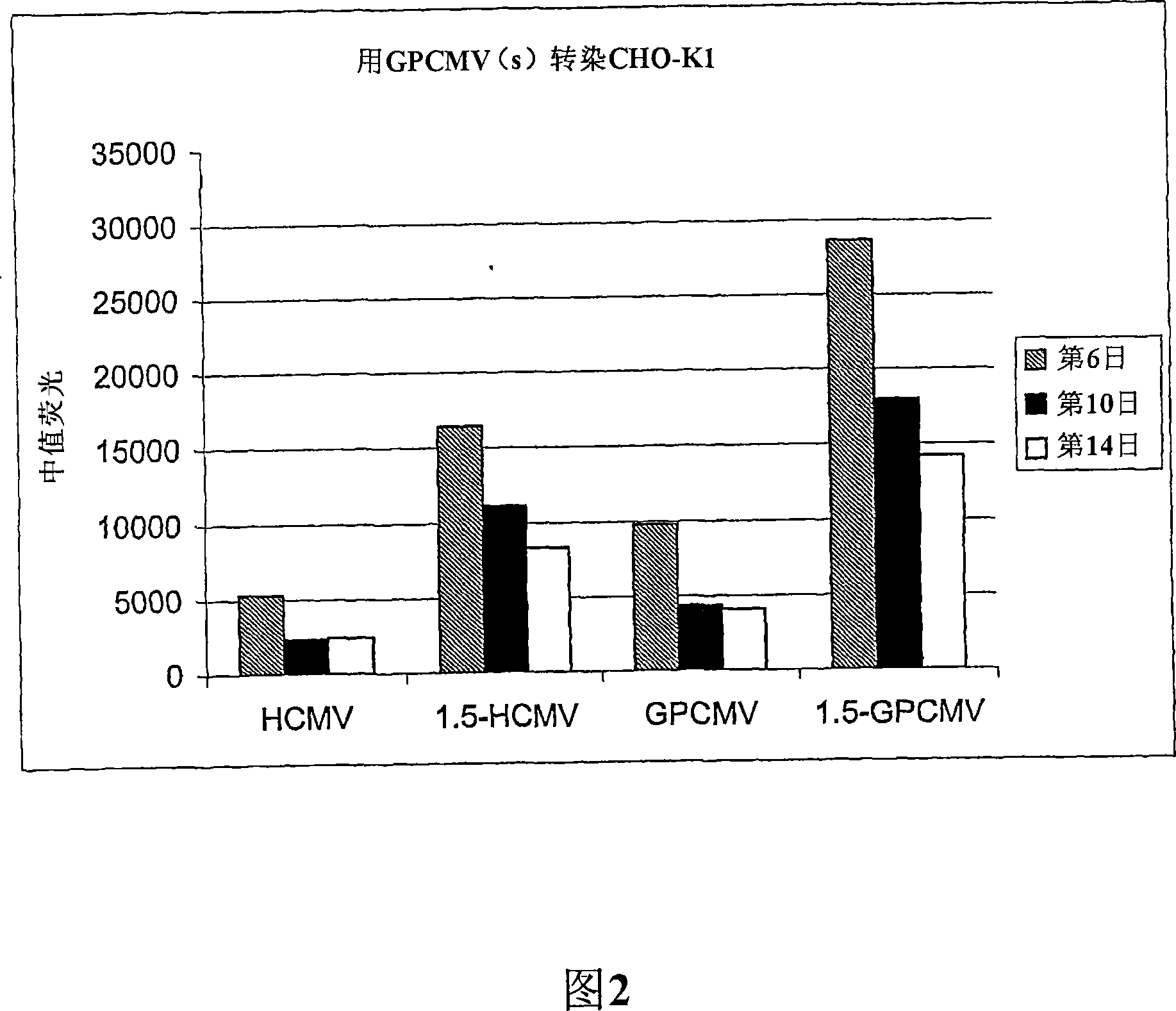
[0077] Generation of stably transfected CHO-K1 cells using vectors including hCMV promoter or gpCMV promoter
[0078] Plasmid constructs were prepared as follows. Ampicillin resistance gene by PCR from pBluescript (Stratagene) isolation, introducing Nru I sites at each end of primers 5'-TGTCGCGAGTCTGACAGTTACCAATGCTTAATC3' (SEQ ID NO: 5), 5'-CATCGCGAGCACTTTTCGGGGAAAT GTGTGCGC-3' (SEQ ID NO: 6). The PCR product was inserted into the Pvu II site of pMae II (Nucleic Acids Research 2001 29:E26) to generate pCA1. The following oligonucleotides
[0079] 1.5′-TCGAAGTTTAAACATTTAAATCTAGAAGCTTAT-3′
[0080] (SEQ ID NO: 7)
[0081] 2.5′-CCGGTATCGATAAGCTTCTAGATTTAAATGTTTAAACT-3′
[0082] (SEQ ID NO: 8)
[0083] 3.5'-CGATACCGGTGGCGCGCCAATTGTTAATTAAGATCTGG-3'
[0084] (SEQ ID NO: 9)
[0085] 4.5'-CCCATTGGGCCAGATCTTAATTAACAATTGGCGCGCCA-3'
[0086] (SEQ ID NO: 10)
[0087] 5.5'-CCCAATGGGCCGTACGAATTCCTTAGGCTCGAG-3'
[0088] (SEQ ID NO: 11)
[0089] 6.5'-GGCCCTCGAGCCTAAGGAATTCGTACG...
Embodiment 2
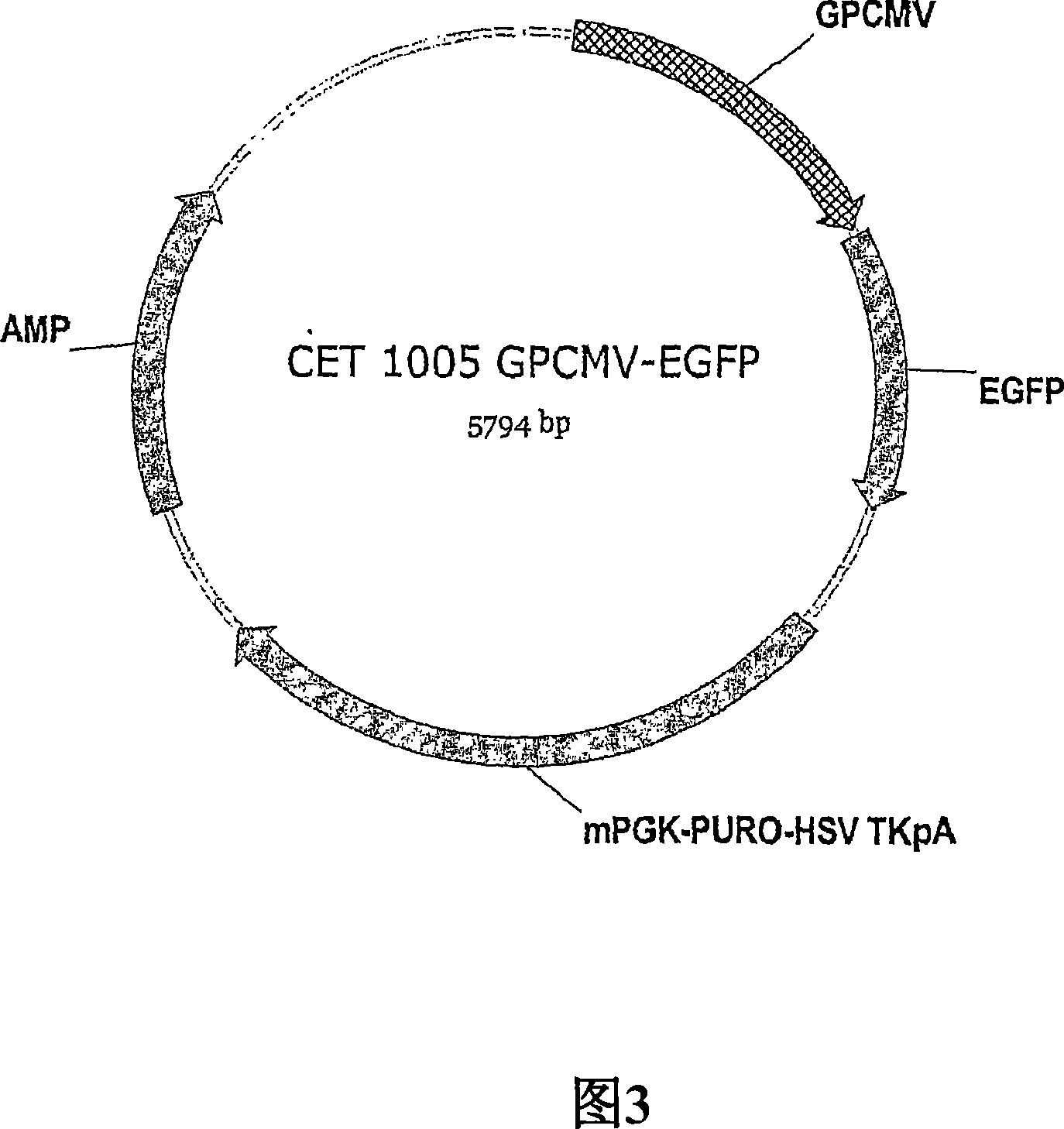
[0095] HEK293 cells were cultured in Dulbecco's Eagle medium (DMEM; Sigma, UK) supplemented with 10% fetal bovine serum and 5 U / ml penicillin and streptomycin mixture (Sigma, UK). For stable transfection, HEK293 cells were treated with 1×10 6 Cells / well density were seeded in 6-well plates and then incubated at 37°C in 5% CO 2 Incubate for 24 hours in the incubator. Cells were then transfected with 4 μg of the indicated plasmids (pCET1005-EGFP or pCET1005-gpCMV-EGFP) (linearized with Pci I) with 10 μl Lipofectamine 2000 (Invitrogen, UK).
[0096] DNA and Lipofectamine 2000 were each diluted in 250 μl of OptiMEM I (Gibco, UK), incubated at room temperature for 5 minutes, mixed and then incubated for another 20 minutes. The growth medium of the cells was replaced with 1 ml OptiMEM I supplemented with 15% FCS, followed by the DNA / Lipofectamine 2000 mixture. Cells were incubated at 37°C in 5% CO before adding 3.5 ml of OptiMEMI supplemented with 10% FCS 2 Incubate for 5 hours ...
Embodiment 3
[0098] CHO-K1 cells were cultured as described in Example 1. Inoculate 1.5×10 cells 24 hours before transferring to 12 wells 5CHO-K1 cells. After 24 hours, cells were transfected with 1 μg luciferase reporter plasmid (phCMV-Luc or pgpCMV-Luc) with 1.5 μl FUGENE (Roche, UK). For this, FUGENE and DNA were diluted separately into Opti-MEM I (Invitrogen), mixed, and incubated for 30 minutes at room temperature before adding to the cells. Luciferase expression was analyzed after 24 hours using a Berthold luminometer (Berthold, Wildbad, Germany). Typically, cell lysis and luciferase reporter assays were performed as previously described (Lipinski et al., Gene Therapy, 2001(8):274-281). Transfections were performed in triplicate and the mean and standard deviation for each representative experiment are shown (Figure 7). It is clear that the luciferase activity of the gpCMV vector is at least 2-fold higher than that of the hCMV plasmid.
[0099] The plasmid hCMV-Luc has been desc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com