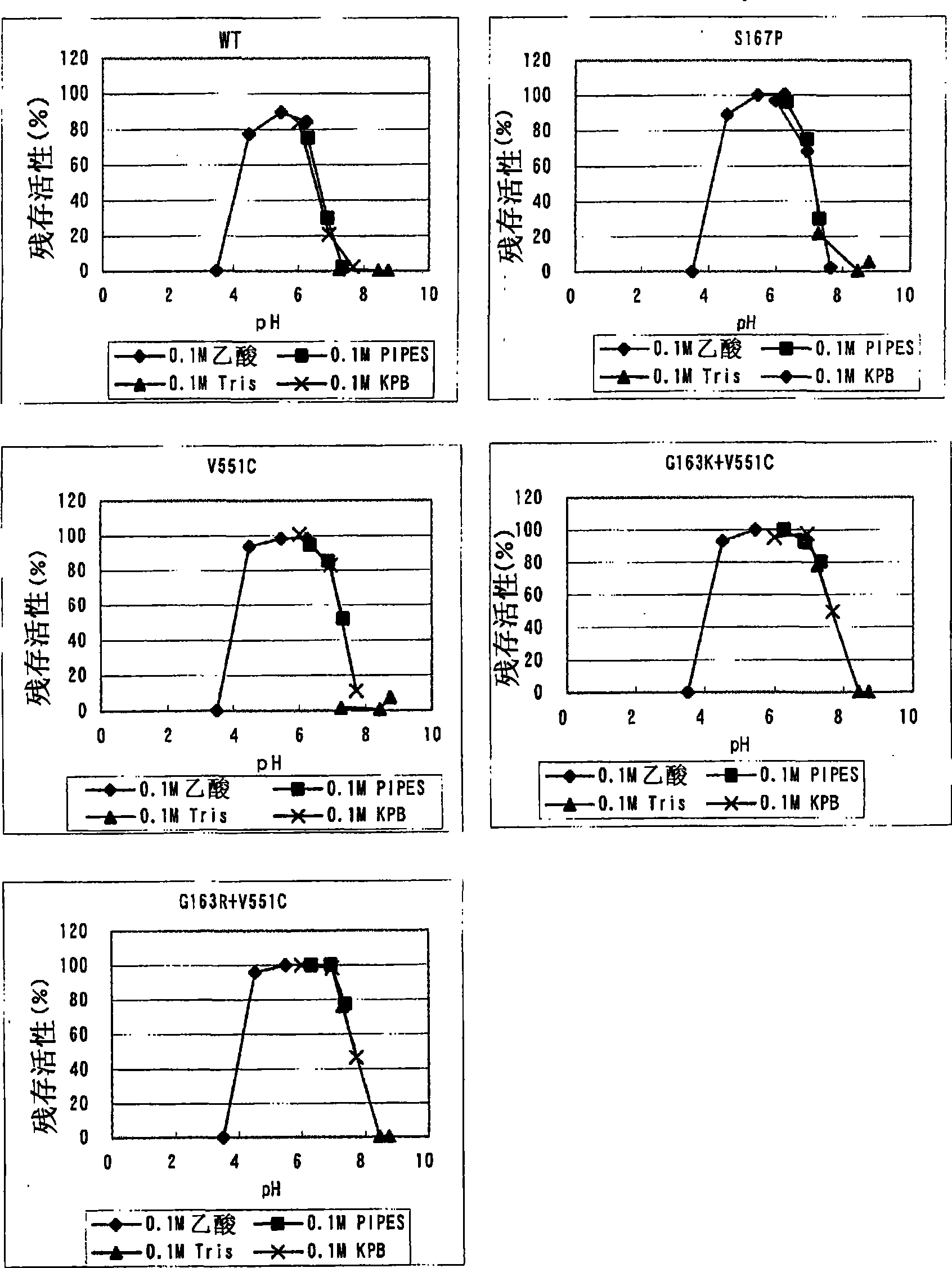
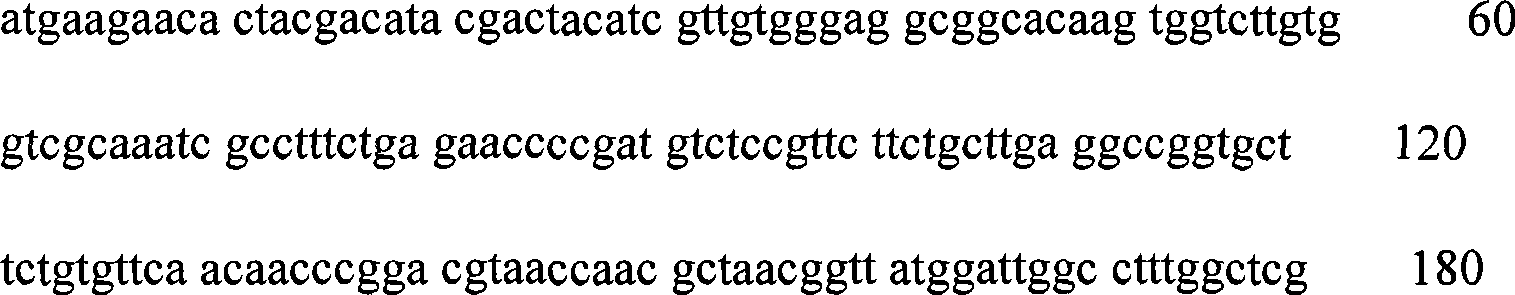Modified flavine-adenine-dinucleotide-dependent glucose dehydrogenase
A glucose dehydrogenase and flavin adenine technology, applied in the field of modified glucose dehydrogenase, can solve the problem of destroying the accuracy of the measured value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0165]In order to obtain the GDH gene derived from Aspergillus oryzae, attempts were made to purify GDH from the culture supernatant of Aspergillus oryzae TI strain in our company using various chromatography, but it was difficult to obtain high-purity GDH, and it had to be abandoned as one of the common methods for gene acquisition. One is the use of partial amino acid sequence cloning. However, we found that Penicillium lilacinoechinulatum NBRC6231 strain produced GDH, and successfully determined a partial amino acid sequence using the purified enzyme. Next, based on the determined amino acid sequence, a part of the GDH gene derived from Penicillium palpinatus NBRC6231 was obtained by using the PCR method, thereby determining the base sequence (1356bp). Finally, based on the base sequence, the Aspergillus oryzae GDH gene was deduced and obtained. This is briefly shown in the following .
[0166]
[0167] [Deduction of Glucose Dehydrogenase (hereinafter also referred to ...
experiment example 2
[0179] [Acquisition of glucose dehydrogenase gene derived from Aspergillus oryzae and introduction into Escherichia coli]
[0180] In order to obtain the AOGDH gene, mRNA was prepared from the cells of Aspergillus oryzae TI strain, and cDNA was synthesized. Two kinds of oligo DNAs shown in SEQ ID NOs: 39 and 40 were synthesized, and AOGDH gene was amplified using KOD Plus DNA polymerase (manufactured by Toyobo Co., Ltd.) using the prepared cDNA as a template. Treat the DNA fragment with restriction endonucleases NdeI and BamHI, and insert the NdeI-BamHI site into pBluescript (corresponding to the LacZ translation initiation codon atg, so that the atg of the Ndel recognition sequence is introduced into the NdeI site in a form corresponding to it) inserted into the recombinant plasmid. Using this recombinant plasmid, Escherichia coli DH5α (manufactured by Toyobo Co., Ltd.) was transformed. Using the transformant, the plasmid was extracted according to a conventional method, an...
experiment example 3
[0182] [Introduction of Aspergillus oryzae-derived glucose dehydrogenase (hereinafter referred to as AOGDH) gene into Escherichia coli]
[0183] In the case of using FAD-GDH after cleavage of the monopeptide as mFAD-GDH, a polypeptide in which only M is added to the N-terminus of mFAD-GDH so that the N-terminus of mFAD-GDH protrudes by 1 amino acid is expressed as S2 .
[0184] In S2, use the oligonucleotide of sequence number 43 as the N-terminal side primer, and carry out PCR in combination with the primer of sequence number 44, and use the same sequence to construct a recombinant plasmid with the DNA sequence (sequence number 1) encoding S2, similarly obtain transformants.
[0185] Wherein, it was confirmed by DNA sequencing that the plasmid having the DNA sequence of modified FAD-GDH has no sequence error.
[0186] The transformant was liquid-cultured for 1 to 2 days in a 10 L fermenter using a TB medium. After the bacterial cells in each culture period were collected, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com