Method for detecting burkholderia cepacia flora on fruits or vegetables and identifying seeds
A Burkholderia cepacia, identification method technology, applied in the field of molecular biology, can solve the problems of long period, cumbersome process, unreliable detection results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Take a little tissue from the junction between disease and health of fruit-rotten apricot in an orchard in Deqing, mash it in 40 μL of sterile water, spread the suspension on a PCAT plate with a sterilized wave stick, and incubate at 28 °C. After 2 days, 23 milky white single colonies were picked from the PCAT plate and purified, numbered Bca01-Bca23.
[0028] Extract 1 to 2 purified bacterial colonies, place them in cell lysate (including 0.25% by weight SDS, 0.05mol / L NaOH), heat at 95°C for 15min, and then add 180 μL bismuth to the cell lysate Distilled in water and stored at -20°C, the resulting solution was used as a DNA template.
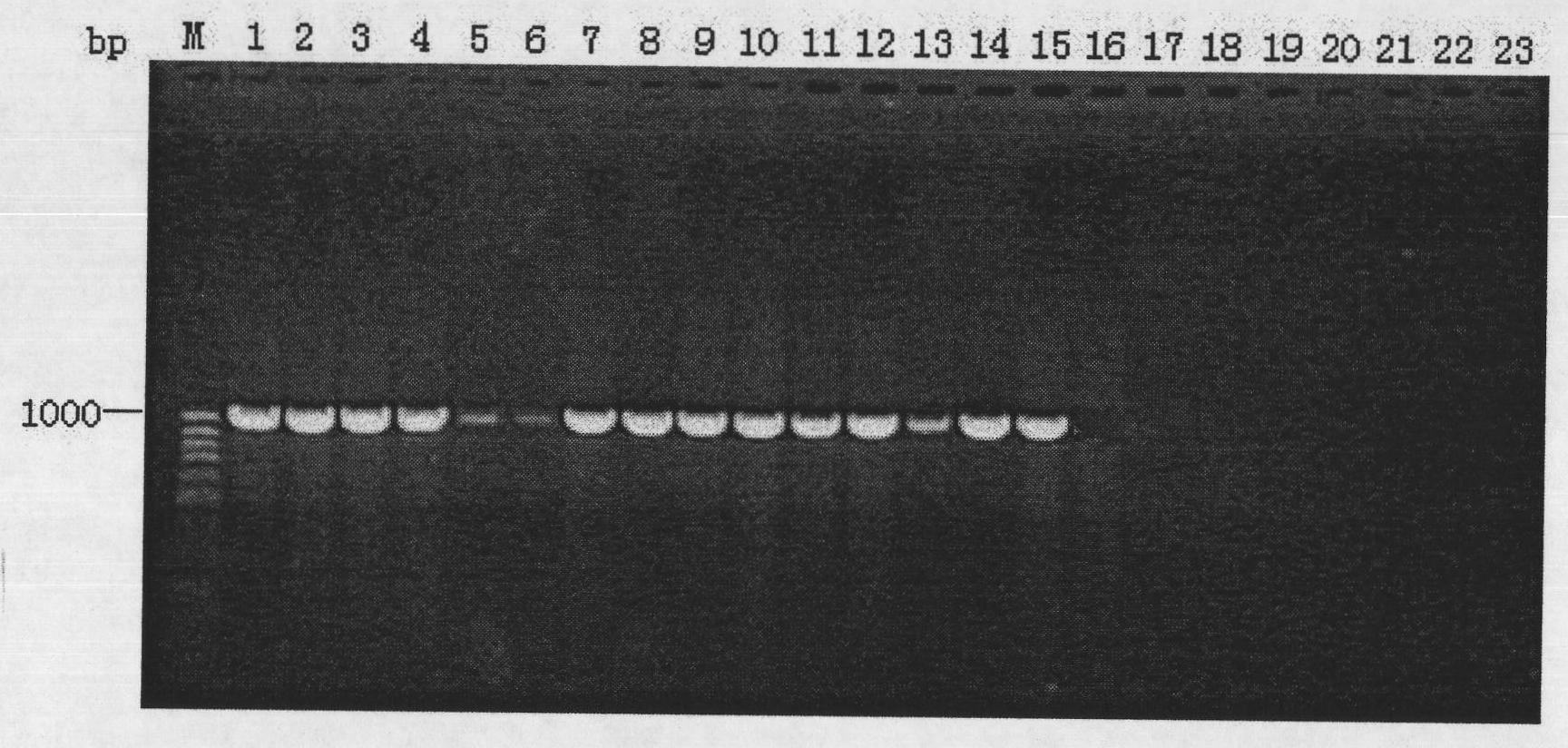
[0029] Using the extracted DNA as a template, PCR amplification was performed using specific primers BCR1 and BCR2 of the recA gene of Burkholderia cepacia.
[0030] Primer BCR1: 5'-TGACCGCCGAGAAGAGCAA-3'
[0031] Primer BCR2: 5'-CTCTTCTTCGTCCATCGCCTC-3'
[0032] The 20μL reaction system for PCR amplification includes:
[0033] Prim...
Embodiment 2
[0039] Digest the PCR products of 15 strains determined to belong to the Burkholderia cepacia group with HaeIII, and the 15 μl enzyme digestion system includes:
[0040] PCR product 10μL
[0041] 10×buffer 1.5μL
[0042] HaeIII endonuclease 1 μL
[0043] wxya 2 O 2.5 μL.
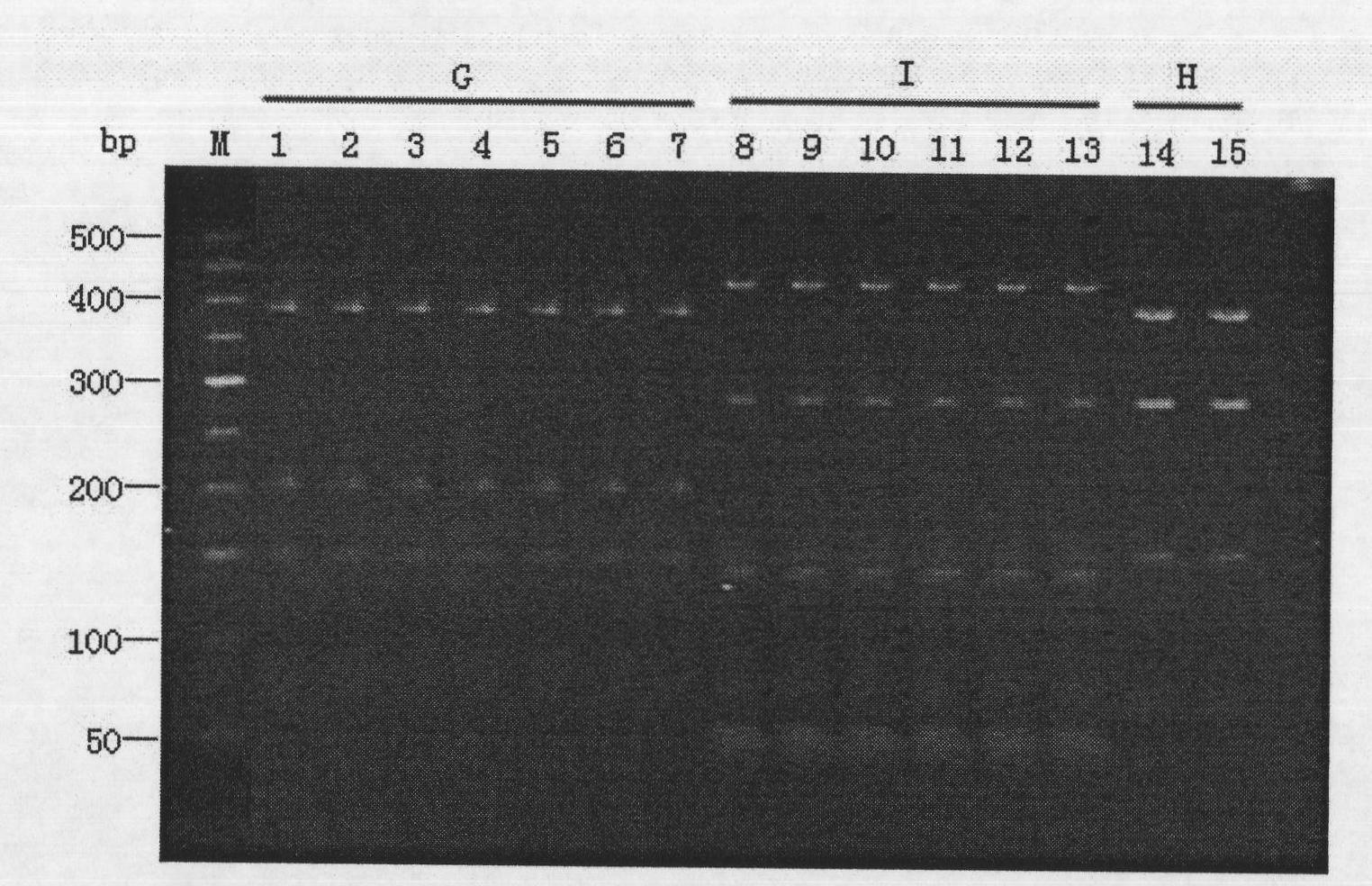
[0044] Digestion as figure 2 As shown, compared with the genotype restriction map reported by Mahenthiralingam et al. (2000) and Dalmastri et al. (2005), it was preliminarily determined that 7 strains Bca01-Bca07 belonged to B. cenocepacia IIIA, and 8 strains Bca08-Bca015 belonged to B. cenocepacia IIIB .
Embodiment 3
[0046] Utilize the specific primers of B.cenocepacia IIIA and IIIB, carry out PCR amplification with the DNA of 7 identifications as B.cenocepacia IIIA and 8 identifications as B.cenocepacia IIIB bacterial strains that embodiment 1 extracts respectively, PCR amplifies The amplification system and reaction conditions are the same as in Example 1.
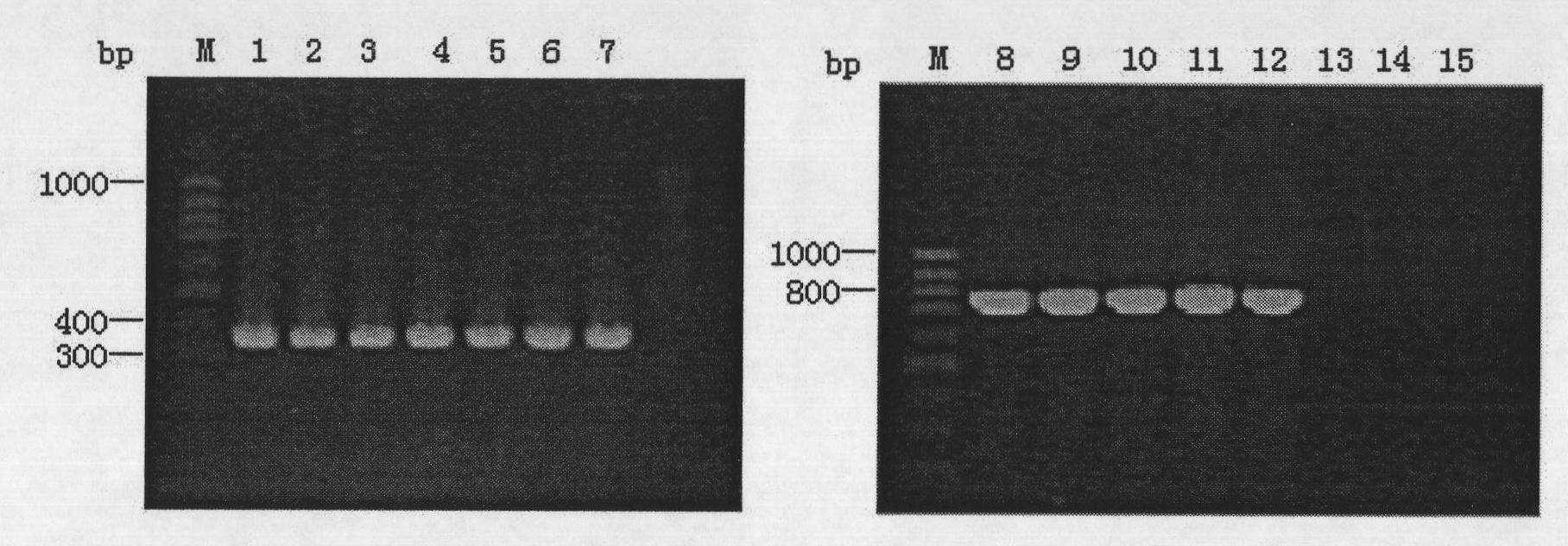
[0047] Take 5 μL of the amplified PCR product and electrophoresis on 1% agarose gel. After electrophoresis, the gel was stained in EB solution for 20 minutes, and then placed on a UV gel imager to observe the results. The results are as follows: image 3 shown. Bc01~Bc07 showed positive amplification to the specific primers of B.cenocepacia IIIA, and Bc08~Bc13 showed positive amplification to the specific primers of B.cenocepacia IIIB, which was consistent with the enzyme digestion results; Bc14 and Bc15 showed positive amplification to B.cenocepacia IIIB The specific primers did not show positive amplification, which was inconsist...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com