Constructing method of nifA gene-introduced rhizobium japonicum genetic engineering bacterial strain HD-SFH-01
A technology of genetically engineered strains, soybean rhizobia, applied in genetic engineering, microorganism-based methods, plant genetic improvement, etc., can solve problems such as small increase in soybean yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
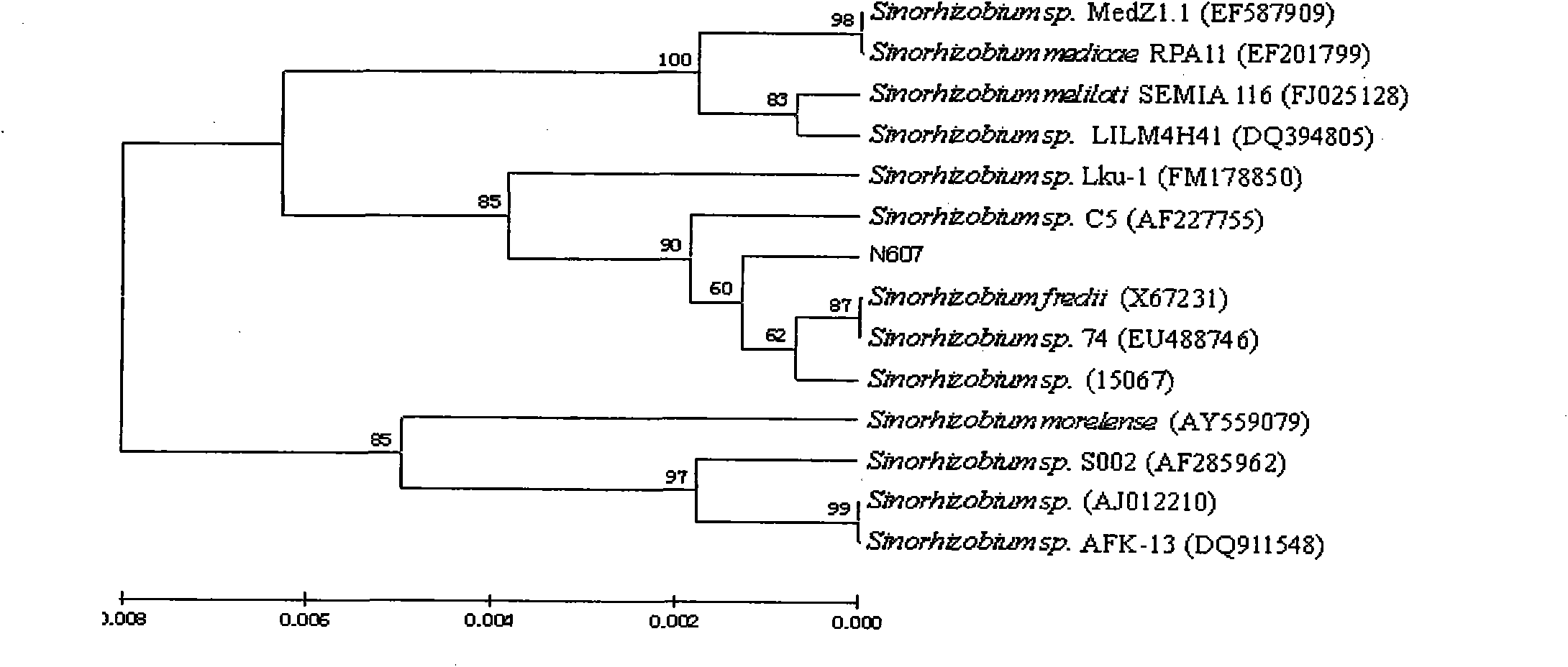
[0010] Specific embodiment one: the construction method of soybean rhizobia genetic engineering strain HD-SFH-01 that imports nifA gene in this embodiment is carried out according to the following steps: 1, fast-growing soybean rhizobium 15067 is activated and expanded, and then the genome is extracted DNA; 2. Using the homologous sequence cloning method, using the fast-growing soybean rhizobium 15067 genomic DNA as a template for PCR amplification to obtain the target gene nifA, and then using TaKaRa Agarose GeL DNA Purification Kit to purify, and then purify the target gene The gene nifA was connected with the TA cloning vector pMD18-T to obtain the connection product; 3. The connection product was transformed into Escherichia coli DH5α and positive clones were screened and identified, and then the plasmid vector pUC19 was extracted by alkaline lysis, and the target gene nifA was combined with Plasmid vector pUC19 was digested by SacI and SalI respectively, and the desired ta...
specific Embodiment approach 2
[0037] Specific embodiment two: the difference between this embodiment and specific embodiment one is that the reaction system of PCR amplification in step two is a 25 μ L reaction system, which consists of the following components:
[0038] Ingredients Dosage
[0039] 400ng / μL fast-growing soybean rhizobia 15067 genomic DNA 1.0μL
[0040] 0.2 μM PnifA1 1.0 μL
[0041] (5'-CGC GTCGACT TATGACTGAATTATCC-3')
[0042] 0.2 μM PnifA2 1.0 μL
[0043] (5'-CGC GAGCTCA CCGAATTTAGATCTT-3')
[0044] 10×Ex Taq Buffer 2.5μL
[0045] 0.4mMdNTP Mixture 1.5μL
[0046] Taq DNA polymerase 0.5 μL
[0047] Sterile water 17.5μL
[0048] PCR amplification conditions were: denaturation at 94°C for 10 min, denaturation at 94°C for 1 min, annealing at 65°C for 30 s, extension at 72°C for 1 min, a total of 35 cycles, extension at 72°C for 7 min, and incubation at 4°C. Other steps and parameters are the same as those in Embodiment 1.
specific Embodiment approach 3
[0049] Embodiment 3: This embodiment differs from Embodiment 1 to Embodiment 2 in that the specific operation steps of TaKaRa Agarose GeL DNA Purification Kit in step 2 refer to the instruction manual. Other steps and parameters are the same as one of the specific implementation modes 1 to 2.
[0050] In this embodiment, TaKaRa Agarose GeL DNA Purification Kit is Takara Bio Agarose GeL DNA Purification Kit.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com