Method for efficiently expressing DNA polymerase by using pichia pastoris
A high-efficiency expression technology of Pichia pastoris, which is applied in the direction of recombinant DNA technology, botany equipment and methods, biochemical equipment and methods, etc., can solve the problems of reducing enzyme activity, difficult purification, high cost, etc., and achieves improved fidelity and operation The effect of cumbersome steps and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
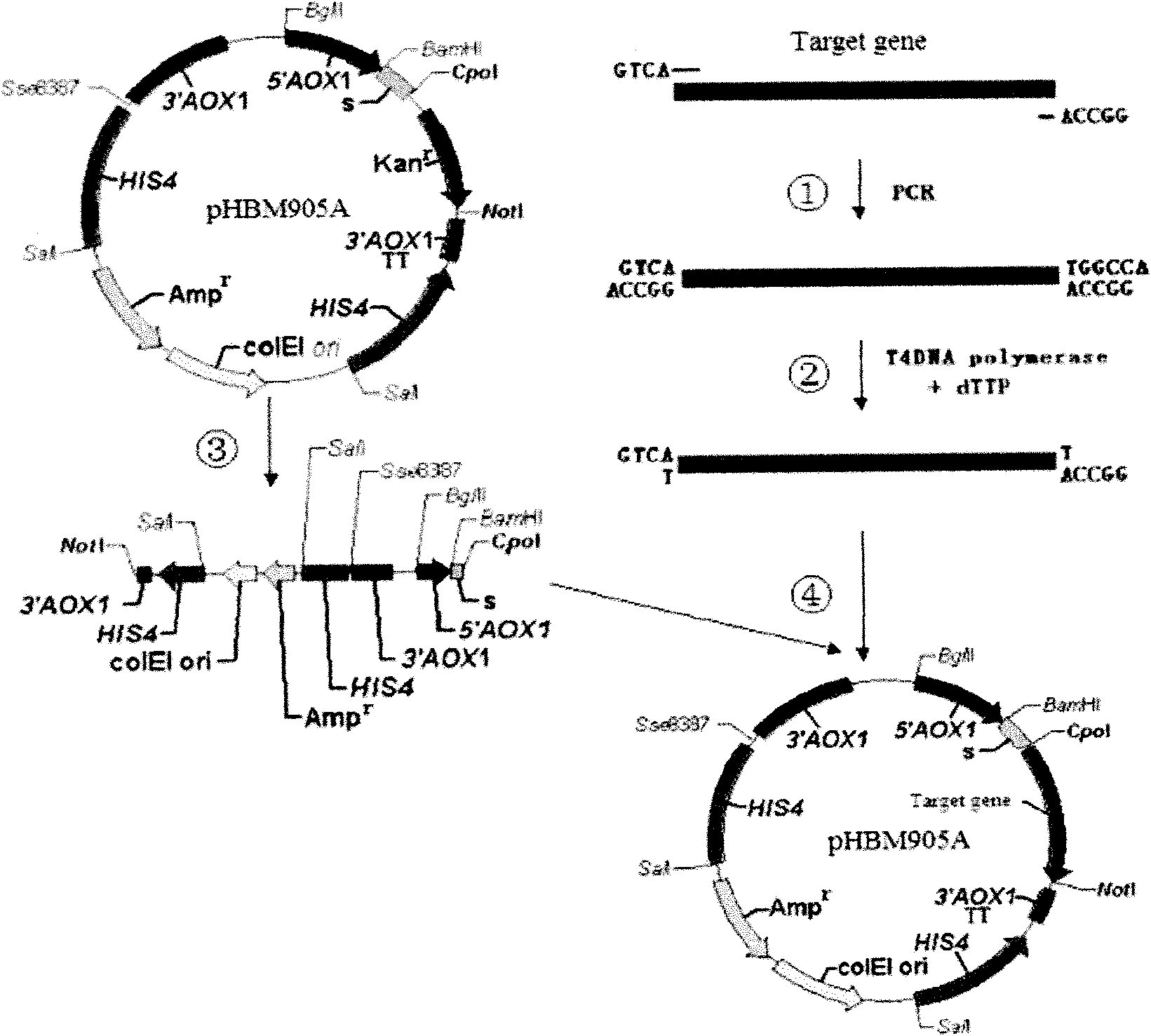
[0027] Using the method of the present invention to express in yeast the DNA polymerase gene derived from the Thermococcus kodakarensis strain synthesized by our laboratory, which is collectively referred to as RKOD hereinafter. Firstly, the pHBM905A plasmid is sequentially digested with restriction endonucleases Cop I and Not I to generate a vector with two fixed base single-stranded cohesive ends, and the vector is recovered by agarose gel. Secondly, according to the known gene sequence, use GeneTool software to design RKOD-905a F: 5′ GTCA ATGATCCTCGACACTGACTACATAACCG 3′ and RKOD-905a R : 5′ GGCCA TATTTTTTCTGTTTTTCCAGCATCTGC 3′ two primers were used for PCR amplification with primerstar DNA polymerase to obtain the RKOD (2.5kb) gene fragment, which was recovered from the agarose gel and treated with T4 DNA polymerase at 12°C for 20min in the presence of dTTP , generate cohesive ends that pair with the pHBM905A vector, and recover fragments. Then the vector and the fragme...
Embodiment 2
[0029] The DNA polymerase gene (RKOD gene) derived from Thermococcus kodakarensis strain synthesized in our laboratory was expressed by the Escherichia coli expression system, as a comparison of Example 1, to verify the superiority of the method of expressing DNA polymerase by yeast. First, according to the known gene sequence, use GENETOOL software to design primers (forward primer: 5'ATGATCCTCGACACTGACTACATAACCG 3'; reverse primer: 5'TTATTTTTTTCTGTTTTTCCAGCATCTGC 3') to amplify the RKOD gene fragment, and agarose gel electrophoresis detects clear bands After banding, the fragment was recovered from an agarose gel. Then the pET-30a plasmid was digested with the restriction endonuclease EcoRV at 37°C for 2 hours to form a linear blunt-ended vector. Then the vector and the fragment were ligated by Solution I enzyme at 16°C for 2 hours, then transformed into Escherichia coli DH10β competent cells, the transformants were screened on the Kanamycin LB plate, and the recombinant was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com