Neutral phytase CP53 from rumen and gene and application thereof
A phytase and neutral technology, applied in the field of genetic engineering, can solve problems such as no neutral phytase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Extraction of Rumen Environment Metagenome
[0053] The metagenomic DNA of goat and cattle rumen fluid microorganisms were extracted by direct lysis method. The specific steps are as follows: 1. Take 50 mL of rumen juice, filter with gauze to remove undigested grass roots and large particles, and wash some microorganisms adsorbed in the grass roots with an appropriate amount of sterilized phosphate buffer; 2. Transfer the supernatant to multiple new In a 50mL centrifuge tube, add twice the volume of preheated lysis buffer, bathe in 70°C water for 2 hours, and shake gently every 10 minutes to mix; 3. After taking it out, cool to room temperature, centrifuge at 5,000rpm for 10 minutes, discard the precipitate, and repeat centrifugation Once; 4. Add 0.7 times the volume of isopropanol to the supernatant, mix gently, place on ice for 30 minutes, centrifuge at 10,000rpm for 30 minutes, wash the precipitate with 70% ethanol once, vacuum dry for 10 minutes, dissolve ...
Embodiment 2
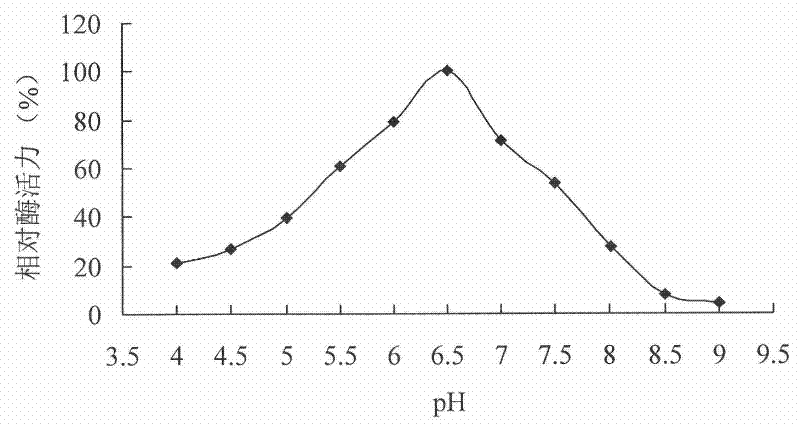
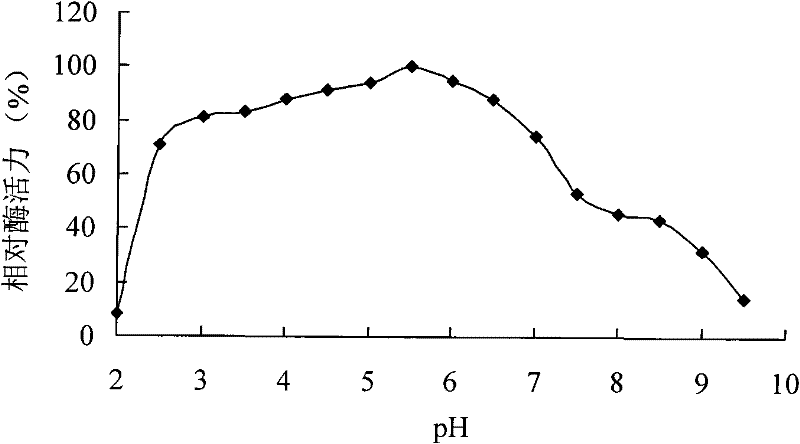
[0054] Example 2 Cloning of phytase-encoding gene cp53
[0055] According to the conservation of CP phytase gene ([V / M / I]-D-L-R-[Q / K / E]-E-[S / T]-H and W-L-H-F-H-C-[X]-[A / E]-G -[X]-G-R-T-T) sequence design and synthesis of degenerate primers P1, P2
[0056] P1: 5'-GTGGACCTRCGRSARGARWCICA-3';
[0057] P2: 5'-GTCCGACCATTGCCTGCYTCRCARTGRAMRTGIADCCA-3').
[0058] PCR amplification was performed using purified rumen fluid metagenomic DNA as a template. Drop-down PCR reaction parameters are: denaturation at 94°C for 5min; denaturation at 94°C for 30sec, annealing at 58-52°C for 30sec, extension at 72°C for 1min, 10 cycles (1°C drop for each cycle), denaturation at 94°C for 30sec, annealing at 52°C for 30sec , 72°C for 1min, 30 cycles, 72°C for 10min. A fragment of about 400bp was obtained, which was recovered and connected with the pEASY-T3 vector and sent to Sanbo Biotechnology Co., Ltd. for sequencing.
[0059] According to the 400bp nucleotide sequence obtained by sequencing, ...
Embodiment 3
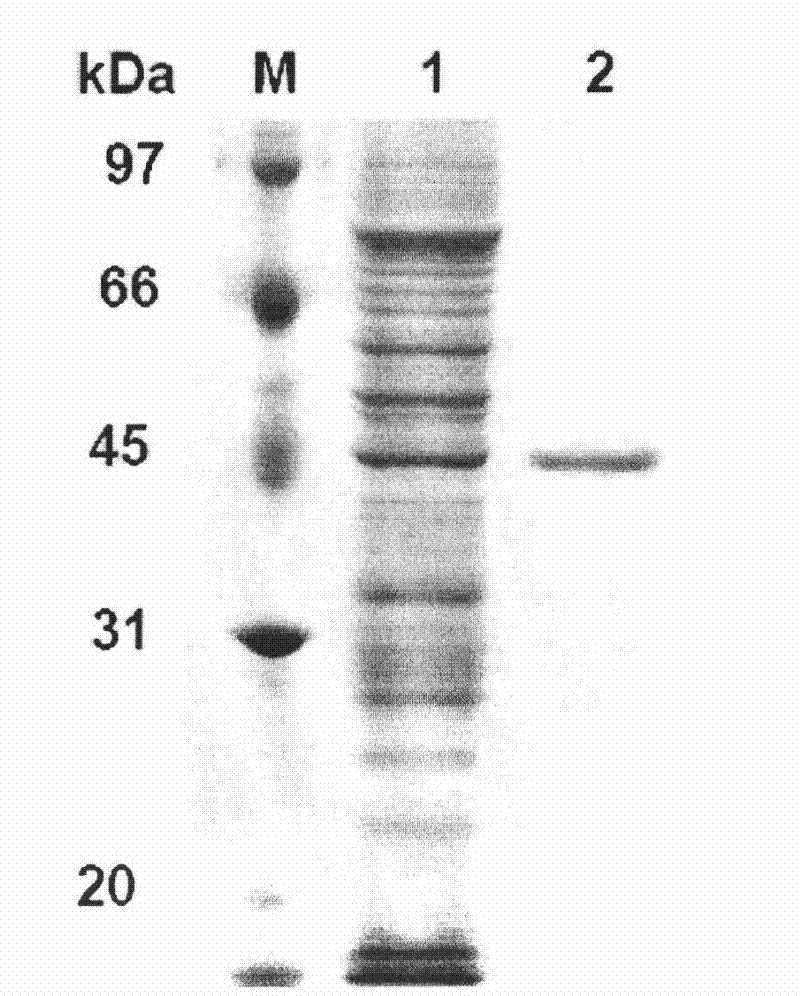
[0063] Example 3 Preparation of recombinant phytase.
[0064] The expression vector pET22b(+) was double digested (NcoRI+XhoI), and the gene cp53 encoding phytase was double digested (NcoRI+XhoI) to cut out the gene fragment encoding mature phytase and the expression vector pET22b( +) connection to obtain recombinant plasmid pET22b-cp53 containing phytase gene cp53 and transform Escherichia coli BL21 (DE3) to obtain recombinant Escherichia coli strain BL21 cp53.
[0065] The BL21 strain containing the recombinant plasmid was inoculated in 3 mL of LB (100 μg / mL ampicillin) culture solution, and cultured at 37° C. overnight. Inoculate 1% of the bacteria in 20mL LB (with 100μg / mL ampicillin), shake and culture at 37°C for about 2-3h (OD 600 After reaching 0.6-0.8), the inducer IPTG with a final concentration of 0.6mmol / L was added, and the culture was shaken at 180rpm at 20°C for 12h. Centrifuge at 12000rpm for 5min, and collect the medium supernatant.
[0066] After being ind...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com