Method for effectively expressing cationic antibacterial peptides in pichia pastoris
A Pichia pastoris, cationic technology, applied in the field of group engineering, can solve the problems of toxicity of host cells, protease attack, consumption of large manpower and material resources, etc., and achieve the effects of reducing toxic effects, increasing expression, and good application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Cloning and sequence analysis of the acid xylanase gene of embodiment 1
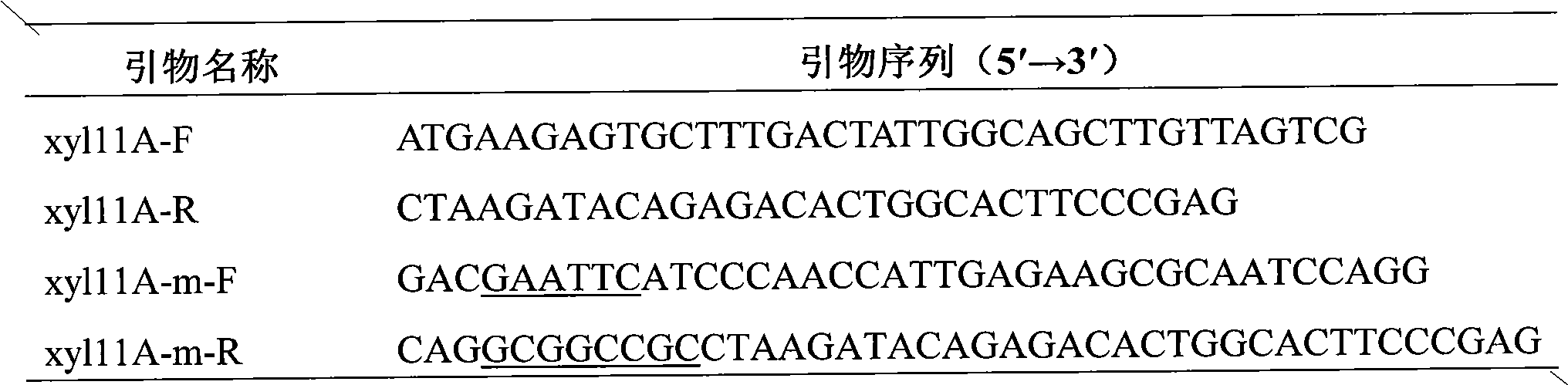
[0021] Total RNA was extracted from Bispora sp.MEY-1 using a total RNA extraction kit for RT-PCR. Use 5 μg of total RNA as the template oligo (dT) as the primer to synthesize the first strand of cDNA, and use the upstream and downstream primers xyl11A-F and xyl11A-R (see Table 1) of acid xylanase XYL11A to obtain xylanase by PCR Full-length fragment of cDNA of gene xyl11A. Cloning the fragment into pEASY-T 3 On the vector, get pEASY-T 3 -xyl11A, after sequencing proved to be correct, it was used as a template to clone the gene sequence of xylanase mature protein.
[0022] Design primers according to the cloned xylanase XYL11A mature protein gene coding sequence: xyl11A-m-F and xyl11A-m-R, and introduce enzyme cutting sites EcoR I and Not I at the end of the primers to sequence the correct pEASY-T 3 The -xyl11A plasmid was used as a template for PCR amplification. The PCR reaction parameters a...
Embodiment 2
[0028] Example 2 Synthesis of the gene of cationic antimicrobial peptide limulusin precursor and KEX2 enzyme cleavage site
[0029] Artificial synthesis of antimicrobial peptide genes
[0030]
[0031] **Note**
[0032] KR: It is the recognition site of yeast's own protease KEX2, which is cut at the rear end of R
[0033] EA: It is the recognition site of yeast's own protease STE13, which is cut at the back end of A
[0034] KWCFRVCYRGICYRRCKR: is the sequence of the cationic antibacterial peptide limulusin Ta I with antibacterial activity
[0035] GKRNEVRQYRDRGYDVRAIPDETFFTRQDEDEDDDEE: Antimicrobial peptide precursor partial sequence
[0036] According to the codon preference of Pichia pastoris, the sequence modification was carried out without changing its amino acid sequence, and the appearance of AT-rich sequences was avoided as far as possible, and these sequences were related to the stability of mRNA. And design the multiple cloning sites BamH I-Xho I and EcoR I ...
Embodiment 3
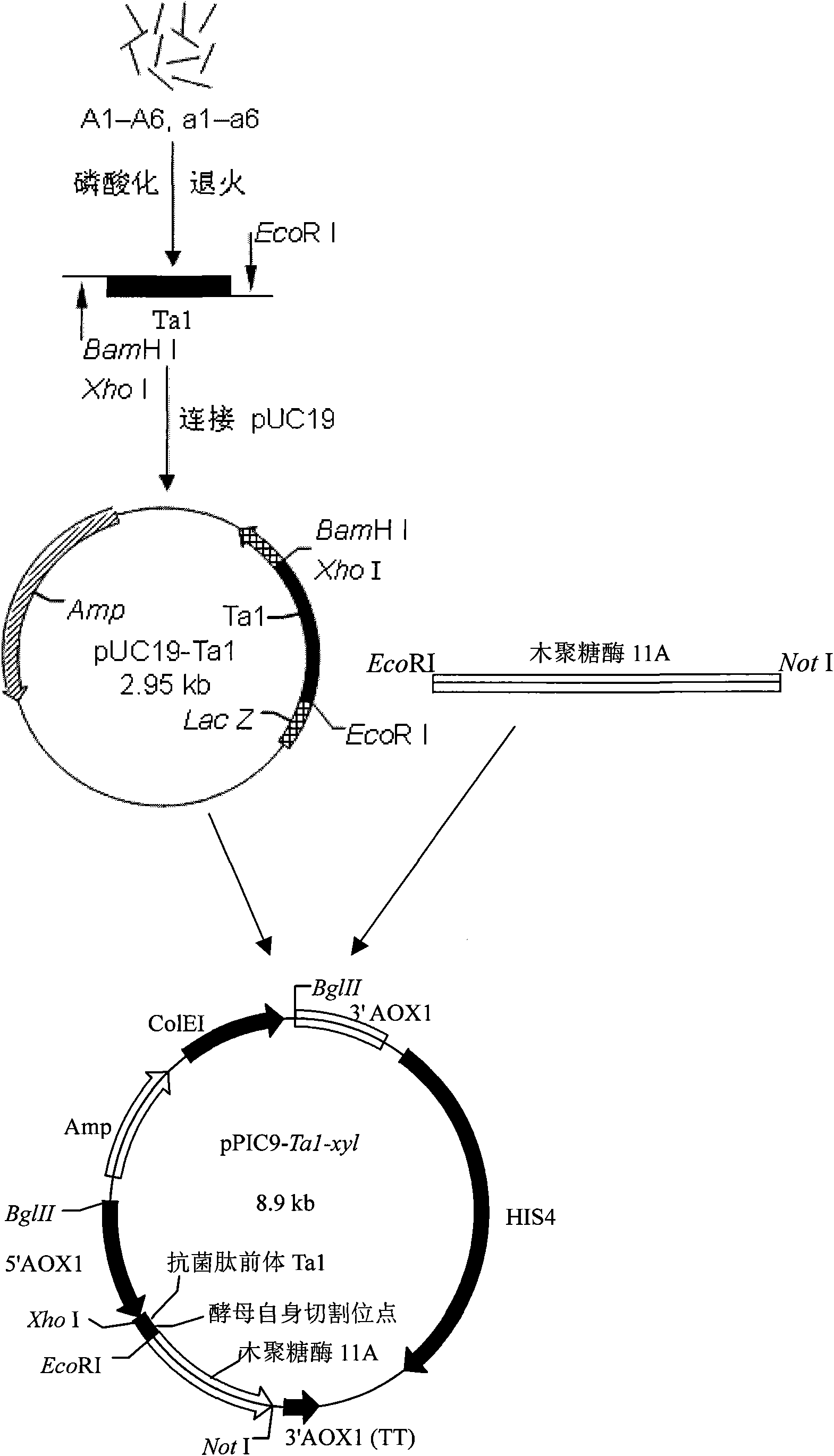
[0057] Embodiment 3 Construction of Fusion Gene Ta1-xyl Expression Plasmid Vector
[0058] First, the gene of the precursor of Tachyplesin I containing the KEX2 restriction site was connected to the Pichia pastoris expression vector pPIC9 to obtain pPIC9-Ta1, that is, pUC19-Ta1 and pPIC9 were first treated with Xho I and EcoR I restriction endonucleases, The vector pPIC9 and Ta1 fragments were recovered after electrophoresis, and pPIC9-Ta1 positive clones were selected after ligation transformation. Using the same method again, acid xylanase xyl11A was connected to pPIC9-Ta1, that is, the acid xylanase xyl11A obtained by EcoR I and Not I restriction endonuclease treatment pPIC9-Ta1 and PCR amplification In the protein region, the vector pPIC9-Ta1 and xyl11A fragments were recovered after electrophoresis, and the fusion expression vector pPIC9-Ta1-xyl was obtained after ligation and transformation. The positive clones of pPIC9-Ta1-xyl were selected by enzyme digestion verificat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com