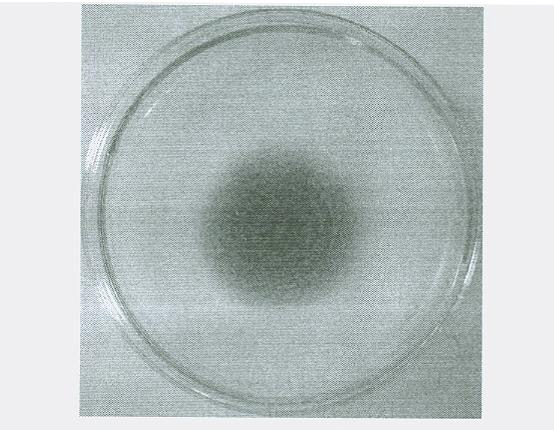
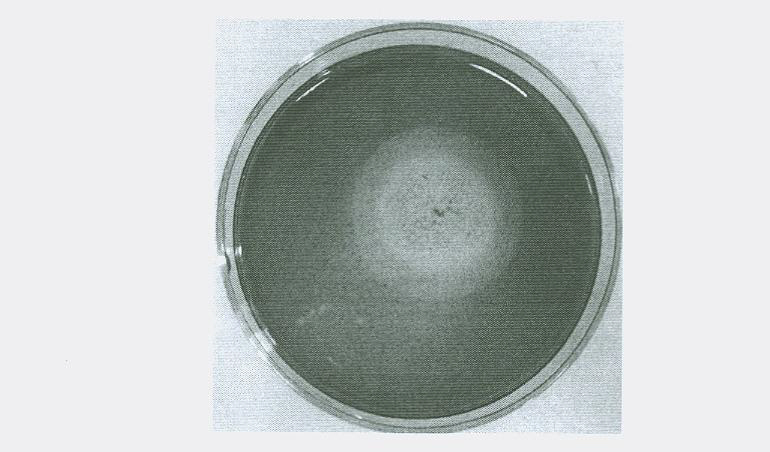
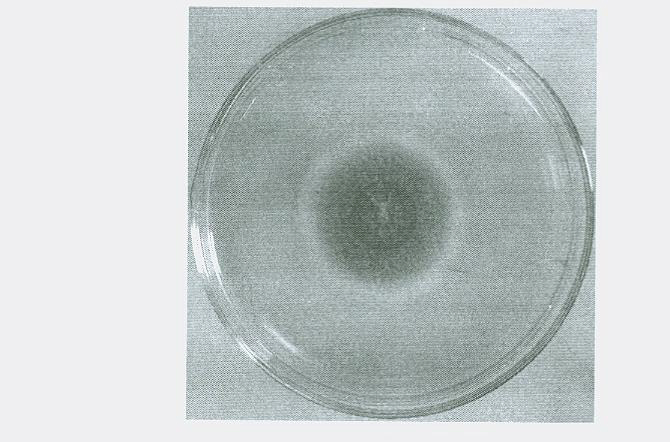
Raw-starch amylase producing penicillium and raw-starch amylase preparation produced thereby
The technology of raw starch and enzyme preparation is applied in the application field of liquid raw amylase preparation and raw tapioca starch simultaneous saccharification and fermentation to produce alcohol, which can solve the problems of lack of market competitiveness and high production cost of fuel alcohol, avoid high-temperature cooking, Solve high cost, cost saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] In embodiment 1, embodiment 2, embodiment 3, raw amylase activity assay adopts 3,5-dinitrosalicylic acid (3,5-dinitrosalicylate, DNS) method (Miller GL.Use of dinitrosalicyclic acidreagent for determination of reducing sugar.Analytical Chemistry1959, 31:426-428), the specific steps are as follows:
[0042] 1. Dissolve glucose in sterile distilled water to make glucose standard solutions of different concentrations; add 1mL DNS to 500μL glucose standard solution, develop color in a boiling water bath for 5min, cool to room temperature, and measure the light absorption value at 540nm; get light absorption Value and the standard curve of glucose concentration, the function formula of standard curve is y=2.7489x-0.0168 (R 2 =0.9994) (y is the light absorption value, x is the glucose concentration).
[0043] 2. Suspend 2 g of raw cassava powder in 100 mL of disodium hydrogen phosphate-citric acid buffer solution with pH 4.0 to obtain raw cassava powder suspension; add 450 μ...
Embodiment 2
[0060] Embodiment 2, the optimization of the culture condition that Penicillium GXU20 produces raw amylase
[0061] 1. Preparation of spore liquid
[0062] 1. Sterilize the PDA medium at 121°C for 20 minutes.
[0063] 2. Wash the spores of Penicillium GXU20 that have been subcultured and activated for 5-7 days on the PDA plate and wash them with sterile water to make a spore suspension. The concentration of the spores is 1×10 10 individual / mL.
[0064] 2. Optimization of pH value
[0065] 1. Prepare basic fermentation media with different pH values (4.0, 4.5, 5.0, 5.5 or 6.0).
[0066] 2. The spore liquid of Penicillium GXU20 was inoculated into the culture medium in step 1 at an inoculum amount (volume percentage) of 1%, and cultured at 28° C. and 180 rpm for 5 days.
[0067] 3. Collect the crude enzyme solution as the solution to be tested, and measure the activity of raw amylase.
[0068] see results Figure 5 , the optimal culture pH value of Penicillium GXU20 prod...
Embodiment 3
[0102] Embodiment 3, the preparation of liquid raw amylase preparation
[0103] 1. Prepare optimized fermentation medium
[0104] Prepare an optimized fermentation medium with pH 5.0.
[0105] 2. Preparation of spore solution
[0106] (1) Sterilize the PDA medium at 121°C for 20 minutes.
[0107] (2) Wash the spores of Penicillium GXU20 on the PDA plate for passage and activation for 5-7 days to make a spore suspension, and the concentration of the spores is 1×10 10 individual / mL.
[0108] 3. Preparation of liquid raw amylase preparation
[0109] (1) Put 50mL optimized fermentation medium into a 250mL shake flask.
[0110] (2) The spore liquid of Penicillium GXU20 was inoculated into the optimized fermentation medium at an inoculum amount (volume percentage) of 1%, and cultured at 28° C. and 180 rpm for 11 days.
[0111] (3) Centrifuge the culture at 13,000rpm, remove the bacteria, collect the supernatant as the solution to be tested, and carry out the determination of t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Enzyme activity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com