Phytoene desaturase gene of sphingomonas sp. and application thereof
A phytoene, less sheath technology, applied in the application, genetic engineering, plant genetic improvement and other directions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Embodiment 1: Obtaining of DNA sequence of Sphingomonas paucimobilis crtI gene
[0023] The phytoene dehydrogenase protein sequences of 4 species of Erythrobacterlitoralis HTCC2594, Erythrobacter longus, Erythrobacter sp.NAP1, and Erythrobacter sp.SD-21 from the order sphingomonadales belonging to Sphingomonas paucimobilis (data from NCBI protein Database) was compared with ClustalW using the software MEGA4, and it was found that there were protein conservative sequences TFDA(G)GPT, VGAGTHPGAG. According to the above conserved sequence and codon preference query (data from the Japanese kazusa Codon Usage Database), the following degenerate primers were designed to amplify the partial sequence of the crtI gene.
[0024] Upstream primers:
[0025] crtIsense: 5′-ACSTTYGAYGCNGGBCCSACS-3′
[0026] Downstream primers:
[0027] crtIanti: 5′-VCCNGCVCCSGGRTGSGTVCCNGGCVCCNAC-3′
[0028] Genomic DNA extracted from Sphmgomonas paucimobilis ATCC31461 (purchased from ATCC) (using...
Embodiment 2
[0050] Example 2: Construction of a recombinant strain of Sphingomonas paucimobilis lacking in phytoene dehydrogenase
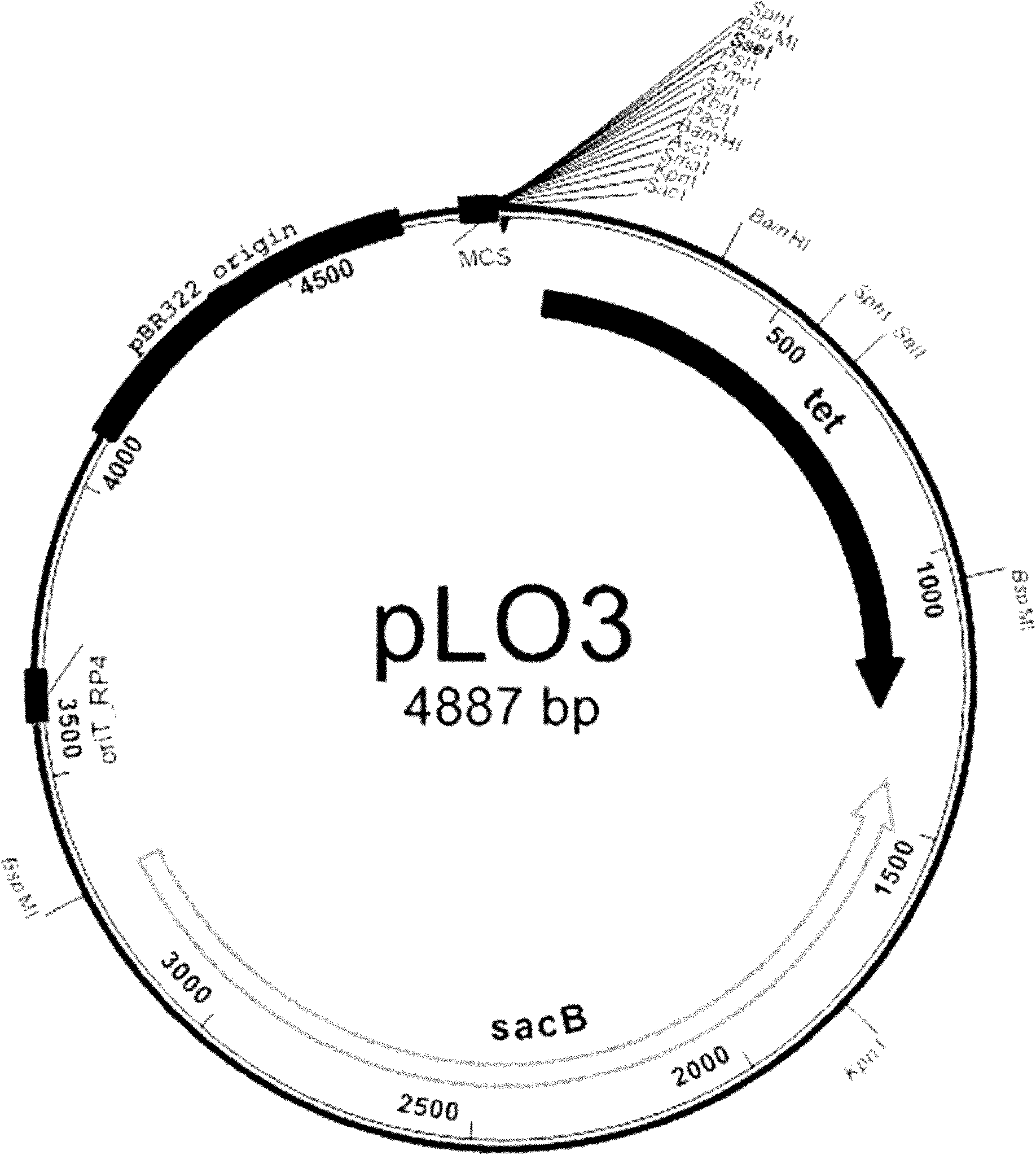
[0051] 1. Construction of gene knockout vector pLO3-ΔI
[0052] Primers were designed based on the obtained DNA sequences. Genomic DNA (using the AxyPrep Bacterial Genomic DNA Miniprep Kit) was extracted from Sphingomonas paucimobilis ATCC31461 (purchased from ATCC) as a template for PCR. I1 and I2 are primers for amplifying the upstream sequence of the crtI gene. The PCR reaction program is: pre-denaturation at 95°C for 5 minutes to enter the cycle process; denaturation at 94°C for 30 sec, annealing at 63°C for 30 sec, extension at 72°C for 1 min, 30 cycles, and finally extension at 72°C 10min. I3 and I4 are primers for amplifying the downstream sequence of the crtI gene. The PCR reaction program is: pre-denaturation at 95°C for 5 minutes to enter the cycle process; denaturation at 94°C for 30 sec, annealing at 64°C for 30 sec, extension at 72°C for 1 min,...
Embodiment 3
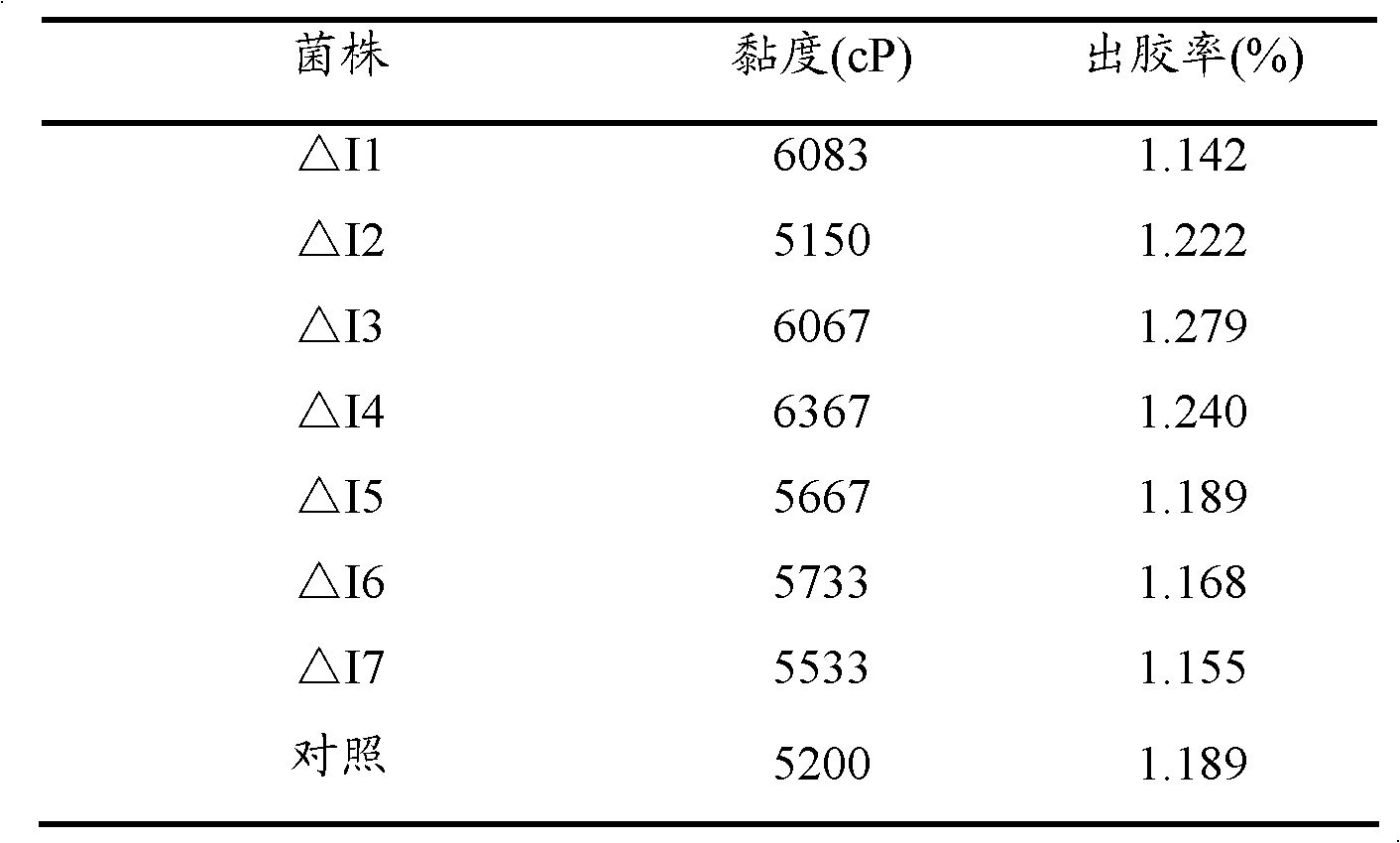
[0076] Example 3: Determination of Gel Production Ability of Sphingomonas paucimobilis Strains Deleted in the crtI Gene
[0077] 1. Wild-type strain (Sphingomonas paucimobilis ATCC 31461) and crtI gene-deleted Sphingomonas paucimobilis (ΔI1~ΔI7) were inoculated on the slant of YM medium, and cultured at 30°C for 72 hours;
[0078] 2. First-level seed culture: respectively insert slant seeds into 50ml first-level seed medium (filled in a 250ml Erlenmeyer flask), and culture at 30°C and 200rpm for 24 hours, which is the first-level seed liquid;
[0079] 3. Secondary seed culture: Inoculate the primary seed solution with 5% volume ratio into 100ml secondary seed culture medium (filled in a 500mL triangular flask), shake and cultivate at 30°C and 200rpm for 12h, which is the secondary seed solution. seed liquid;
[0080] 4. Fermentation: Put the secondary seed liquid into 100mL grade fermentation medium (filled in a 500mL Erlenmeyer flask) with an inoculation amount of 5% volume ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com