Method for detecting gene mutation
A detection method and forward primer technology, which are applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of lack of gene mutation methods, poor detection, and difficult amplification, etc. The effect of detection cost, easy judgment, and short time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] Below in conjunction with embodiment, further illustrate the present invention.
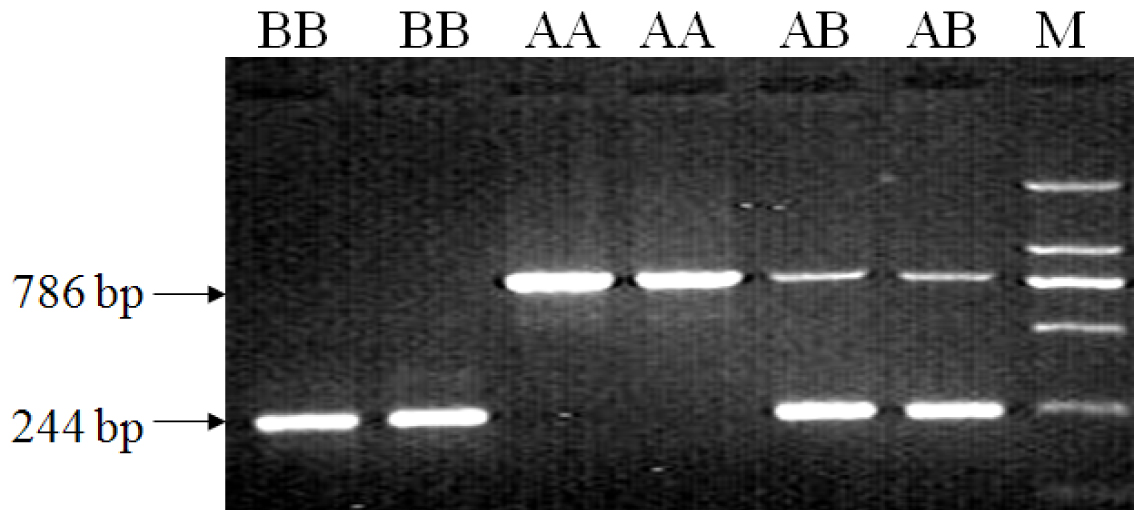
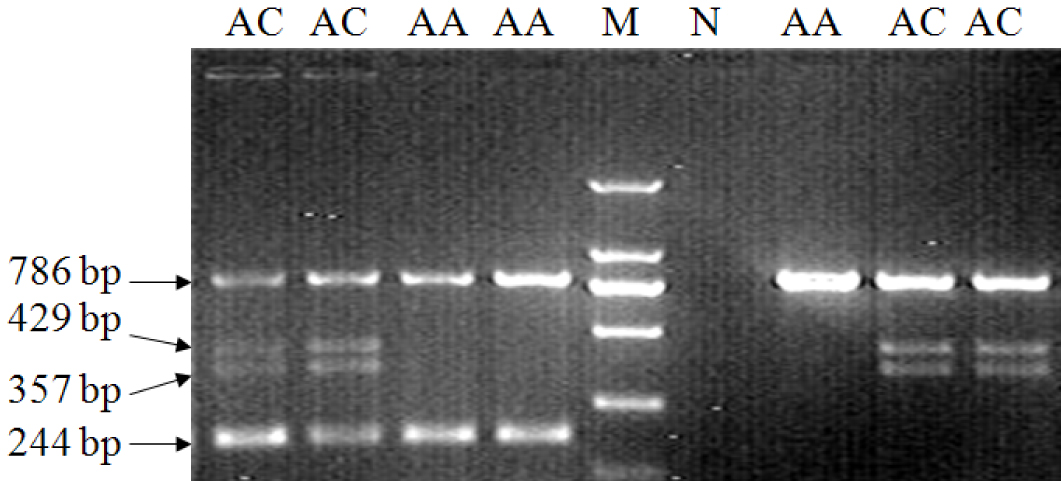
[0027] chicken GHR Establishment of a new method for molecular detection of type I mutations in genes
[0028] (1) Primers:
[0029] take chicken GHR Design primers P1-F / R for the sequence at both ends of the type I mutation deletion fragment on the gene, and design primer P2-F / R according to the sequence at both ends of the type II mutation (reference sequence GeneBank accession number: AC187590), and amplify the obtained sequence and related information such as figure 1 shown. The primer sequences used are as follows:
[0030] P1-F: 5'-GGCAGAAACCCCAAGTATG-3' (SEQ ID NO: 1);
[0031] P1-R: 5'-TCGGGACAGATCAAAGACAAT-3' (SEQ ID NO: 2);
[0032] P2-F: 5'-ATCCTGTTGCTTTCTTATGG-3' (SEQ ID NO: 3);
[0033] P2-R: 5'-CTCAGTCCTGTAGCCCTTG-3' (SEQ ID NO: 4).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com