Expression vector for large green alga bioreactor and transformation method thereof
A technology of bioreactors and expression vectors, which is applied in the construction and transformation of expression vectors, can solve problems such as untrackable detection, and achieve the effect of avoiding false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
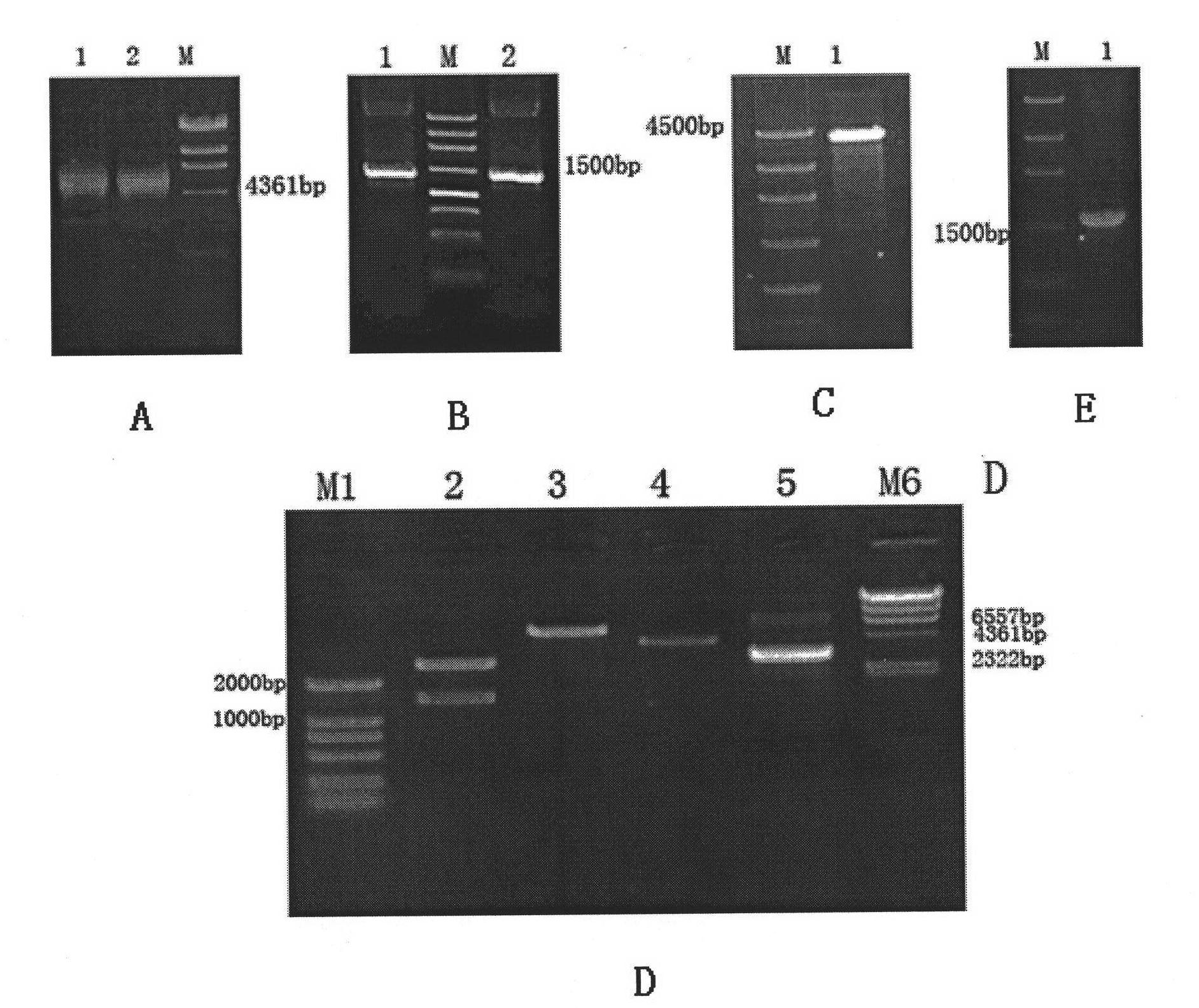
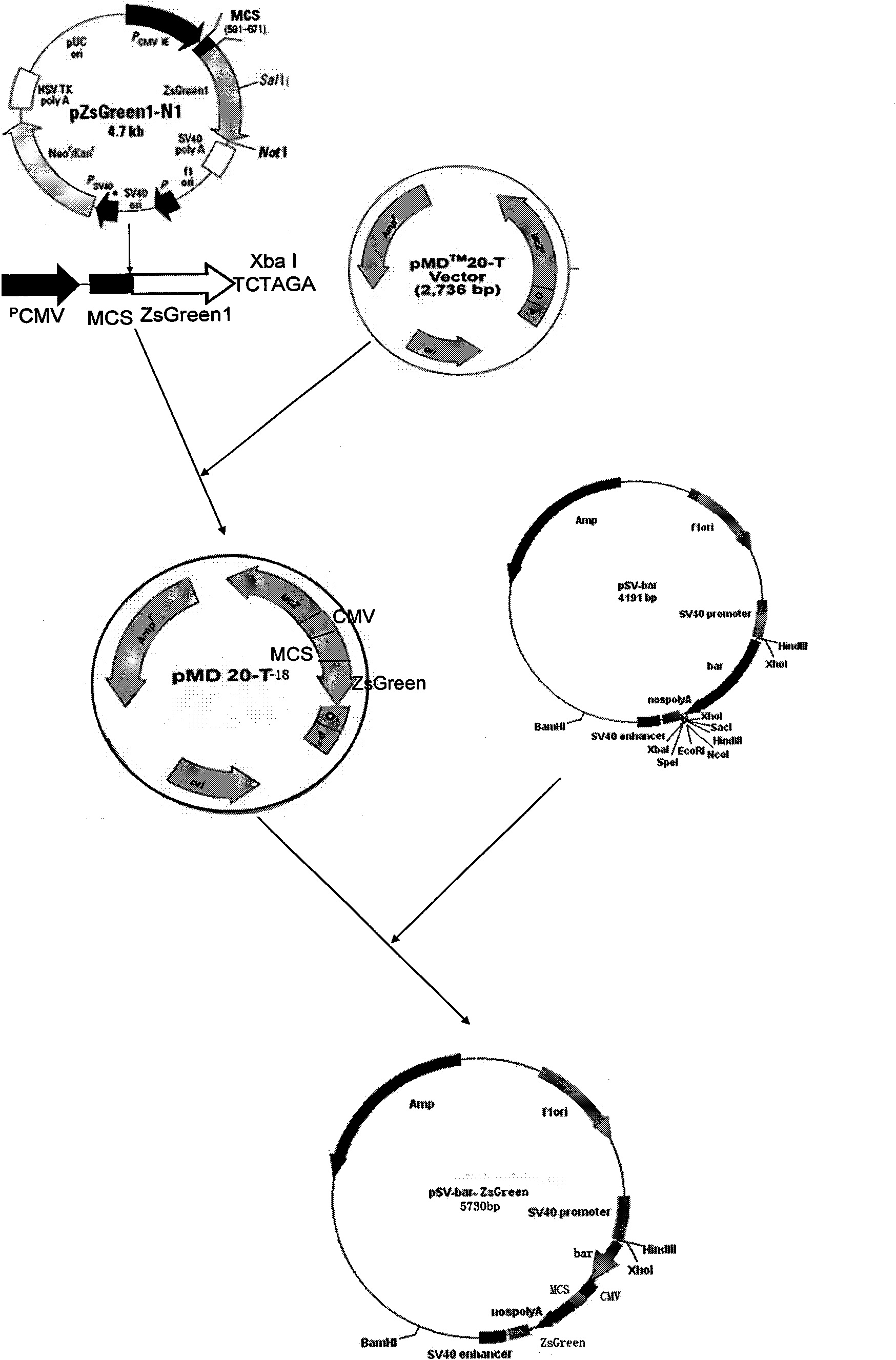
[0043] Example 1 Transformation plasmid vector construction and amplification
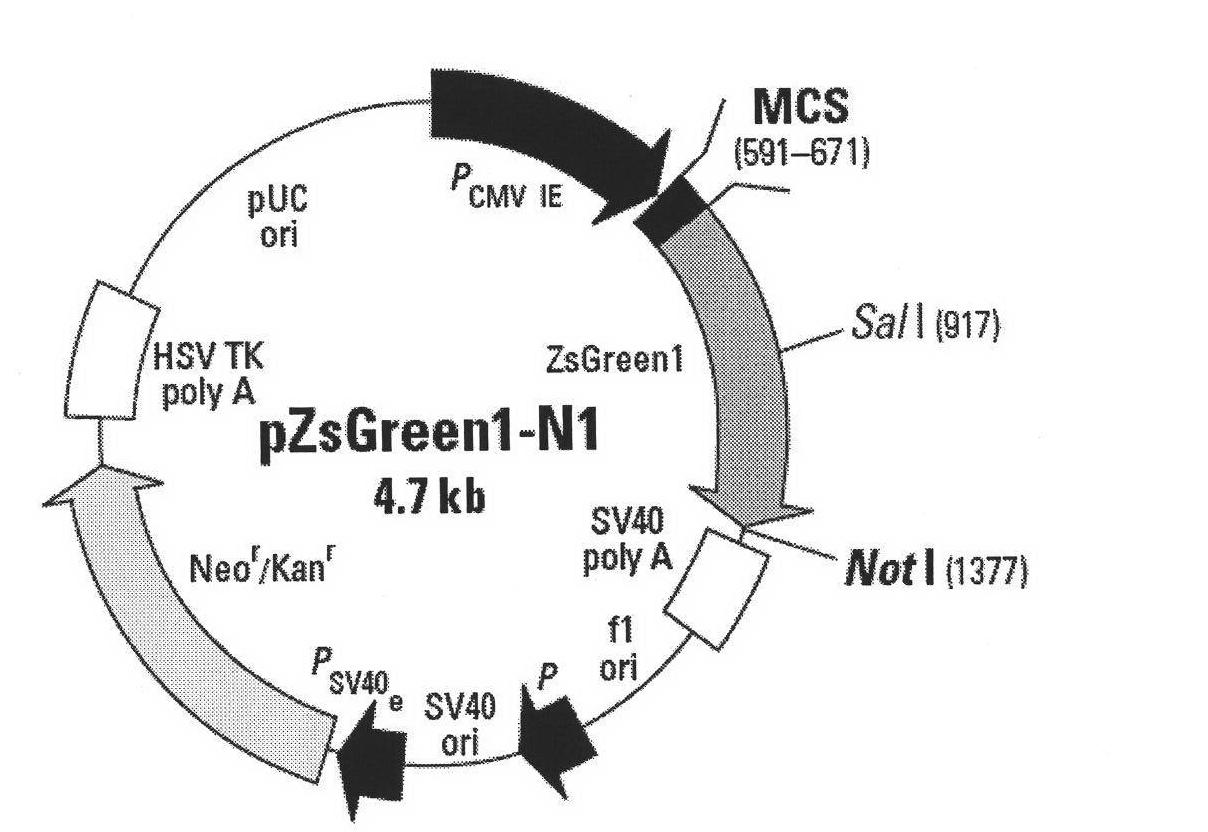
[0044] 1.1 According to the pZsGeenl-N1 expression vector (purchased from Clontech, about 4.7kb), such as figure 1 As indicated, a pair of specific primers was designed. Add Xba I restriction site at the 5' end of upstream primer pZsGeen1 P1 and downstream primer pZsGeen2 P2, its sequence includes CMV immediate earlypromoter, enhancer region, TATA box, start point, start codon, stop codon (about 1500bp).
[0045] The primer sequences are as follows:
[0046] pZsGeen1 P1: 5'-GTTATTAATAGTAATCAATTACGG-3'
[0047] pZsGeen1 P2: 5'-TCTAGATCAGGGCAAGGCGGAGCCGG-3'
[0048] 1.2 The PCR reaction system and conditions for amplification are:
[0049] PCR reaction system (25μl):
[0050] Sterile ultrapure water 17.1μl
[0051] 10×Buffer (including Mg 2+ ) 2.5 μl
[0052] dNTP 1μl
[0053] Primer (10ppm) each 1μl
[0054] Template (0.8μg / μl) 2μl
[0055] Taq enzyme (TAKARA) 0.4μl (2.5U)
[0056] Pre-...
Embodiment 2
[0075] Example 2 Screening of Transformation Selectable Markers of Macroalgae
[0076] Through screening, the herbicide Basta (glufosinate) has a strong killing effect on Enteromorpha spores and seedlings, and Basta at a concentration of 5 μg / ml can completely kill Enteromorpha spores within 3 days, and 12.5 μg / ml It takes about a week under the concentration to kill all the seedlings of Enteromorpha. Subsequent experiments showed that the results were also applicable to other types of Enteromorpha, such as Yuanguan.
Embodiment 3
[0077] Example 3 Optimizing the conditions for the separation and purification of protoplasts from Enteromorpha spp.
[0078] It was found that when protoplasts of Enteromorpha barbadensis and Enteromorpha spp. were obtained by enzymatic method, the optimal enzymatic hydrolysis formula was 2% cellulase mixed with 2% isolated enzyme, the optimum enzymatic hydrolysis pH value was 6.5, and the concentration of osmotic agent mannitol was 0.6 M. Through protoplast culture, it was observed that Enteromorpha cells mainly had three different development modes: single-cell seedlings, cell clusters, and sporangia / gametes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com