Method for reducing dimer of pair of primers with part of sequence being identical
A polymer, pair technology, applied in the field of primer design to reduce the formation of primer dimers, can solve the problems of irregularity, cumbersome, time-consuming target genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment
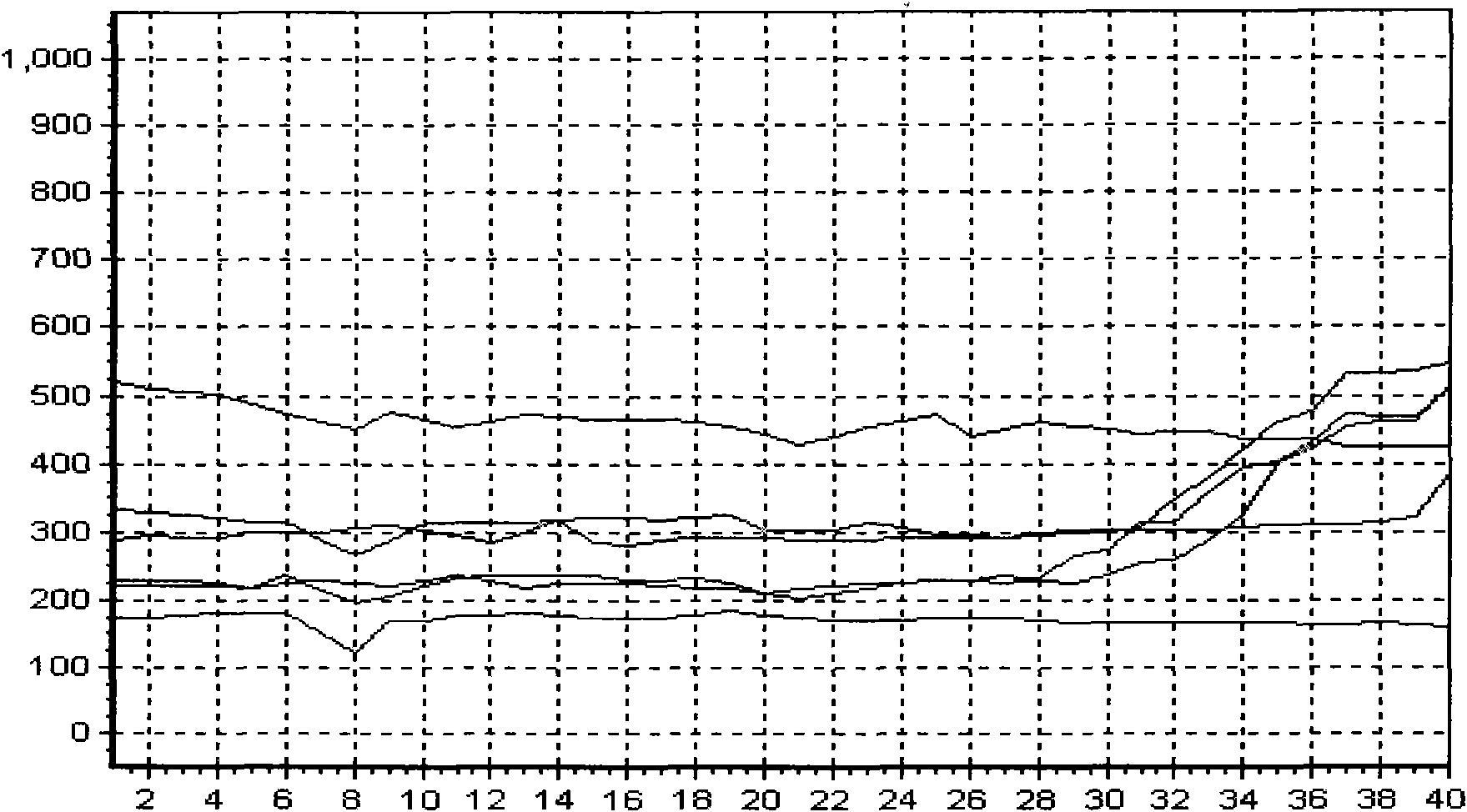
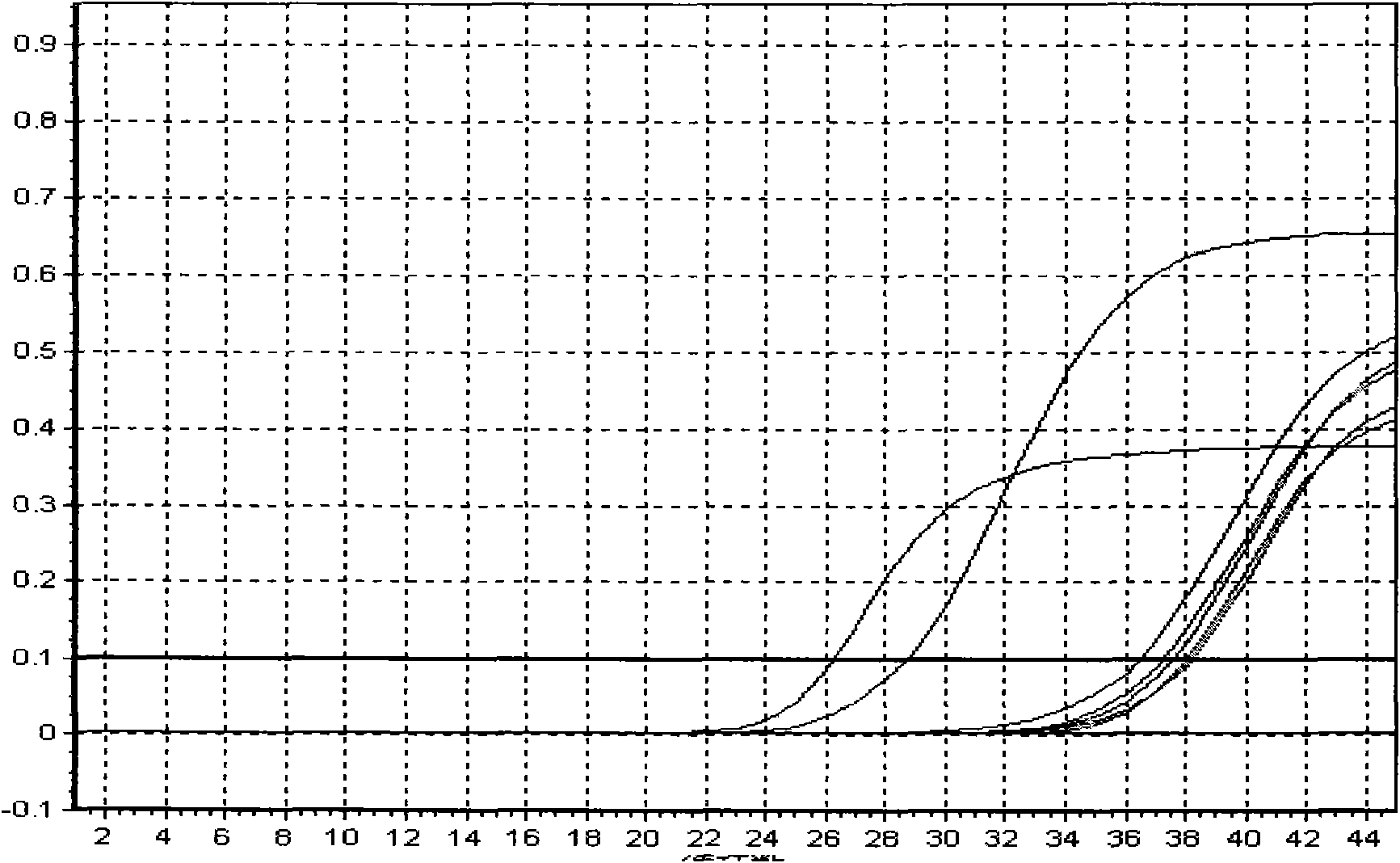
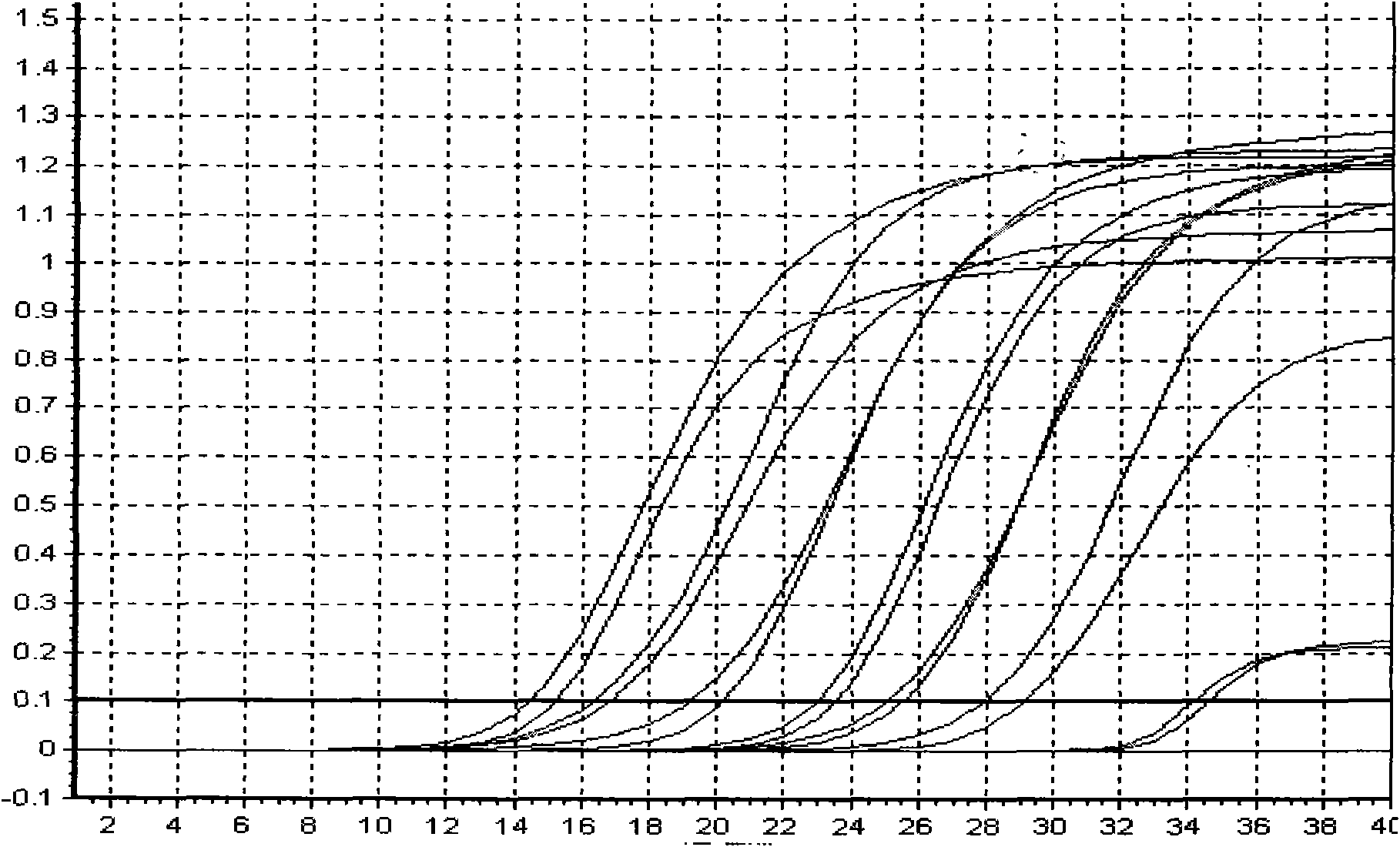
[0089] Specific embodiment: hepatitis B viral load real-time fluorescent PCR detection
[0090] The following examples further illustrate the content of the present invention, but should not be construed as limiting the present invention. Without departing from the spirit and essence of the present invention, the modifications or replacements made to the methods, conditions, steps and applications of the present invention all belong to the scope of the present invention.
[0091] Hepatitis B virus (referred to as hepatitis B) is a worldwide infectious disease caused by hepatitis B virus (Hepatitis B virus, HBV). The infection rate of hepatitis B in our country is very high, which greatly endangers people's health. At present, the detection methods of hepatitis B mainly include enzyme immunoassay, radioimmunoassay, chemiluminescence, immunofluorescence, nucleic acid amplification (PCR) fluorescence quantitative method and so on. Enzyme immunoassay is widely used, but real-tim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com