Rapid construction of Arabidopsis artificial miRNA gene interference vector
A construction method and an Arabidopsis technology are applied in the field of rapid construction of artificial miRNA gene interference vectors in Arabidopsis, which can solve problems such as cumbersome content and so on.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Example 1 Modification of pCAMBIA1300 vector for rapid silencing vector construction
[0096] 1. Preparation of transformation vector
[0097] In order to improve the applicability of this patent, it is particularly emphasized to explain the method of modification of the target vector, so as to provide users of the present invention for reference, and can be used to modify other target vectors when necessary.
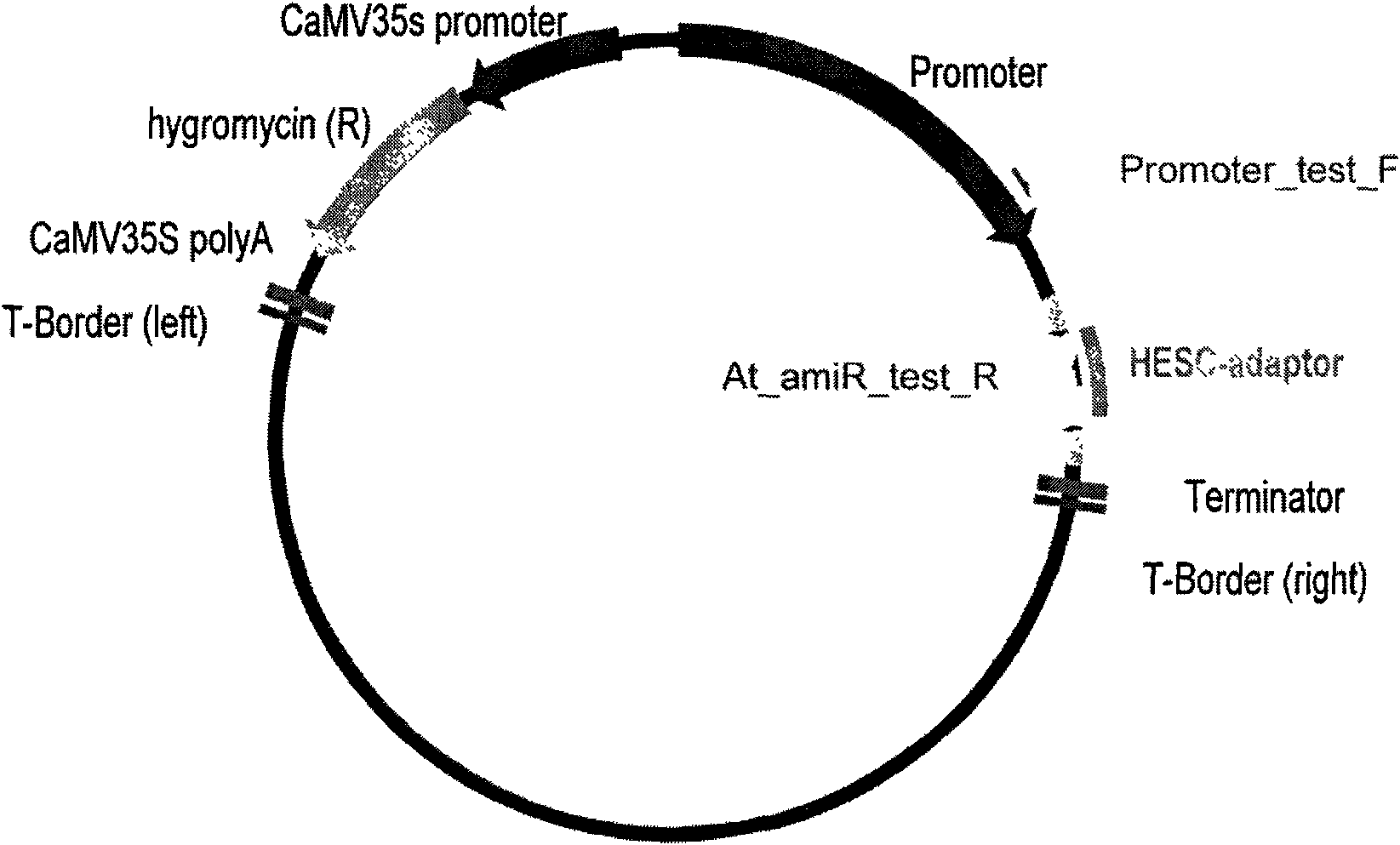
[0098] In order to put the first and last batches into the vector and be able to carry out restriction digestion linearization, we first used the modified pCAMBIA1300 vector as the initial vector, which is connected with the 35s promoter and rbcs terminator, which can be used to start and stop Arabidopsis. This vector is referred to as the 1300SR vector for short.
[0099] First, it was digested with SacI and KpnI endonucleases and digested overnight at 37°C. The digested DNA was electrophoresed on a 1% agarose gel, stained with ethidium bromide, and then cut under a UV ...
Embodiment 2
[0119] Example 2 Rapid construction of 5 Arabidopsis gene silencing vectors using the patented method
[0120] 1. Preparations for the rapid construction of silent vectors
[0121] (1) Plasmid preparation and linearization
[0122] The vector obtained according to Example 1 was amplified by shaking bacteria, and 1300URA plasmid DNA was extracted using a plasmid extraction kit, and digested with AfeI. The digestion system is as follows:
[0123]
[0124] After digestion at 37°C overnight, the digested DNA was electrophoresed on a 1% agarose gel, stained with ethidium bromide and cut under ultraviolet light, and the digested product was recovered using a gel recovery kit. The linearized vector is a universal vector fragment, which can be used for the construction of all rapid silencing vectors and can be stored at -20°C for a long time for later use.
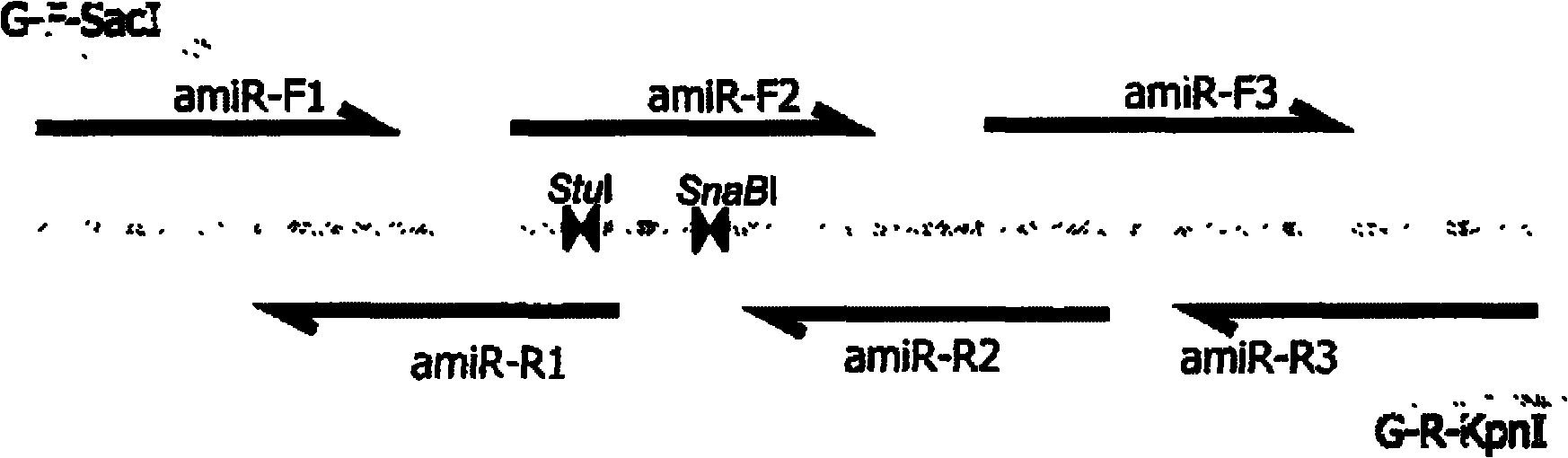
[0125] (2) Synthesis of bridge fragments
[0126] Two primers for bridging fragments were synthesized according to the method of this pate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com