Mutant glucose dehydrogenase
A glucose dehydrogenase and glucose technology, applied in the direction of oxidoreductase, biochemical equipment and methods, applications, etc., can solve problems such as inability to measure blood sugar levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1 Plasmid expressing GDH or CyGDH of Burkholderia cepacia
[0083] Plasmids expressing the α subunit and γ subunit of GDH were prepared as plasmids expressing GDH of Burkholderia cepacia, and plasmids expressing α subunit, β subunit, and γ subunit were prepared as plasmids expressing CyGDH.
[0084] Plasmids expressing the α subunit and γ subunit of GDH
[0085] The plasmid pTrc99A / γ+α described in WO02 / 036779 (corresponding to EP1331272A1, US2004023330A1, CN1484703A) was used as a plasmid expressing the α subunit and the γ subunit. This plasmid is a DNA fragment isolated from the chromosomal DNA of Burkholderia cepacia KS1 strain (FERM BP-7306), which continuously contains the GDH γ subunit structural gene and α subunit structural gene, and is inserted as the cloning site of the vector pTrc99A Point the NcoI / HindIII in the resulting plasmid. The GDHγα gene in this plasmid is controlled by the trc promoter. pTrc99A / γ+α retains the ampicillin resistance gene...
Embodiment 2
[0103] Example 2 Substrate interaction by introducing mutations into the CyGDHα subunit gene site exploration
[0104] (1) Introduce mutations to 326, 365 and 472
[0105] On the GDHα subunit gene contained in pTrc99Aγαβ obtained in Example 1, the 326-position serine residue, the 365-position serine residue, and the 472-position alanine residue of the α subunit encoding the gene Variations are introduced by substitution of other amino acid residues.
[0106] Specifically, using a commercially available site-specific mutation introduction kit (Stratagenc, QuikChangell Site-Directed Mutagenesis Kit), the GDHα subunit gene contained in the plasmids pTrc99A / γ+α and pTrc99Aγαβ described in Example 1 The codon for serine at position 326 (TCG), the codon for serine at position 365 (TCG), and the codon for alanine at position 472 (GCG) were substituted with codons for other amino acids.
[0107] The sequences of the forward primer and the reverse primer used for the above amino ...
Embodiment 3
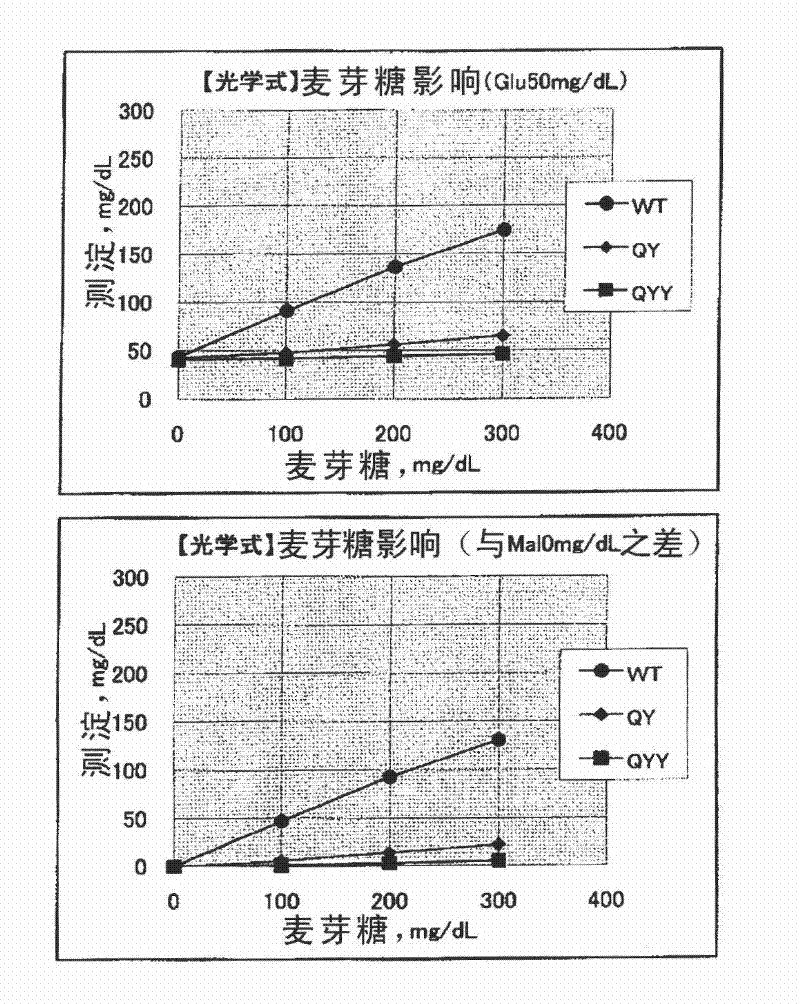
[0121] Example 3 Analysis of the substrate specificity of variant GDH
[0122] Using the mutant GDH expression plasmid obtained in Example 2, a mutant GDH was produced and substrate specificity was studied.
[0123] (1) training
[0124] For the Escherichia coli DH5α strain into which the mutation was introduced, 2 ml each of LB medium (containing 50 μg / ml ampicillin and 30 μg / ml kanamycin) was used and cultured overnight at 37° C. in an L-shaped tube with shaking. These culture solutions were inoculated into a 500 ml slanted flask containing 150 ml of LB medium (containing 50 μg / ml ampicillin and 30 μg / ml kanamycin), and cultured with shaking at 37°C. After 3 hours from the start of the culture, IPTG (isopropyl-β-D-thiogalactopyranoside) was added to a final concentration of 0.1 mM, and cultured for another 2 hours.
[0125] (2) Preparation of crude enzyme samples
[0126] Collect the thalli from the above cultured culture solution, after washing, suspend the thalli in 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com