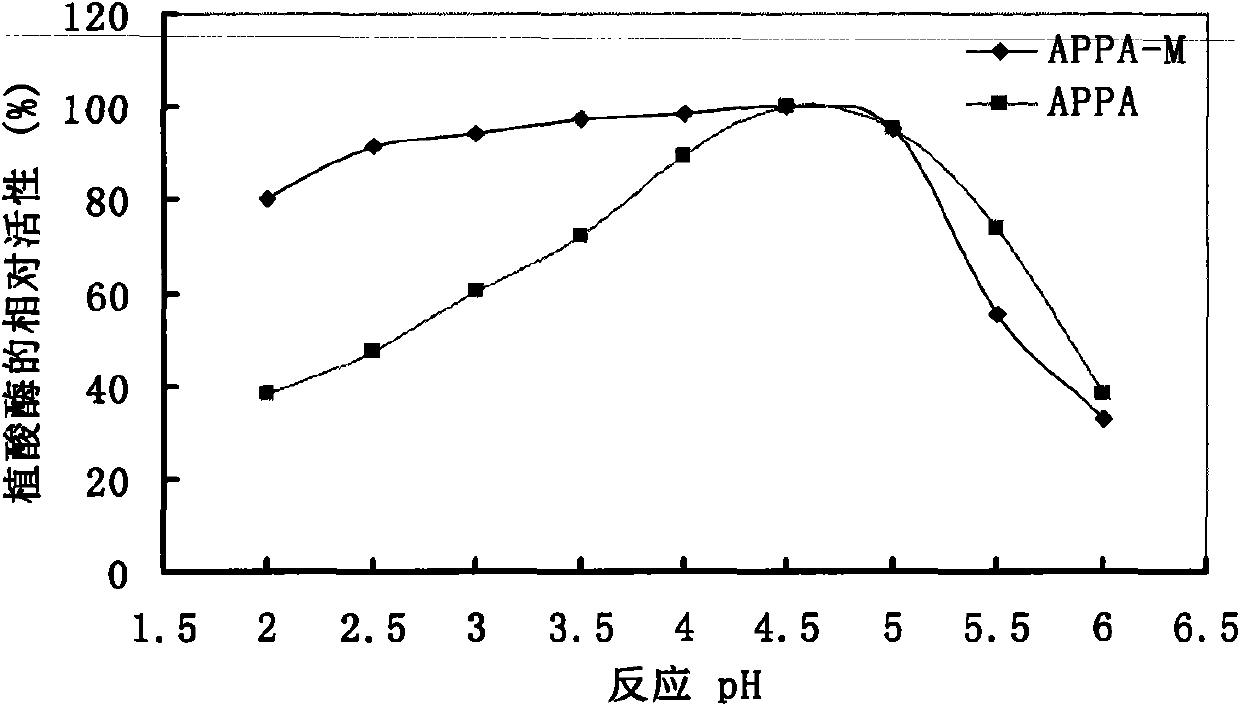
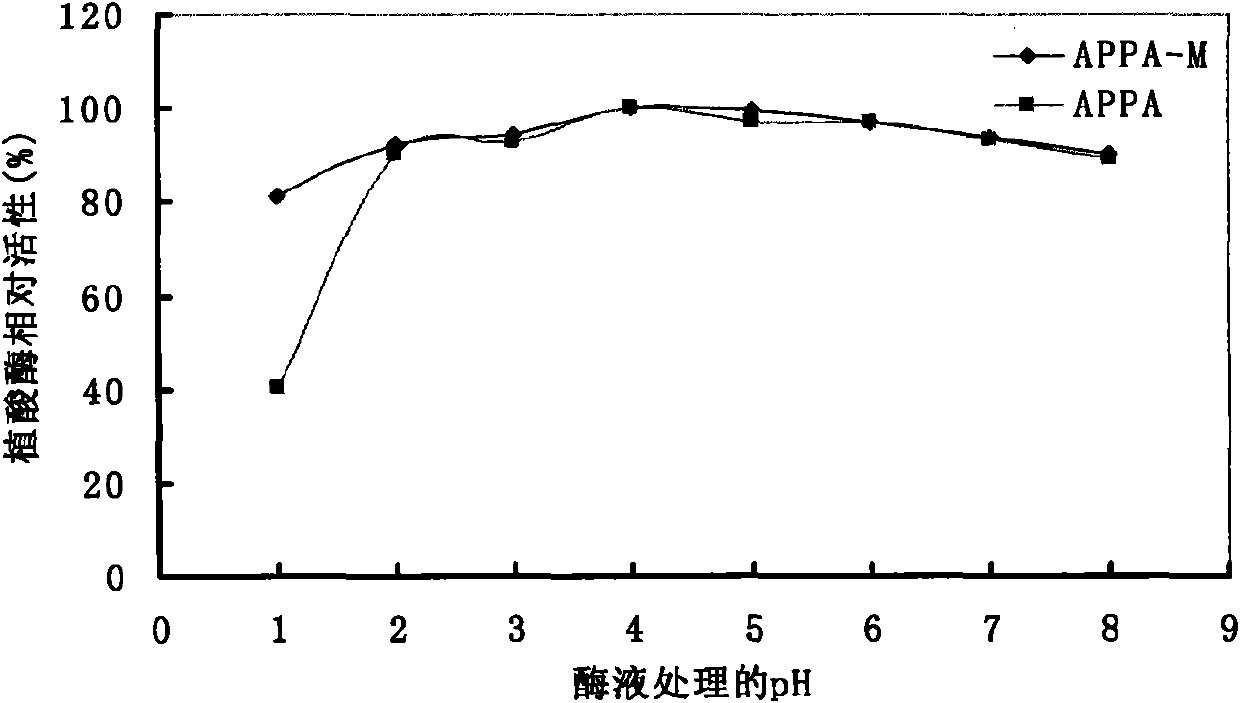
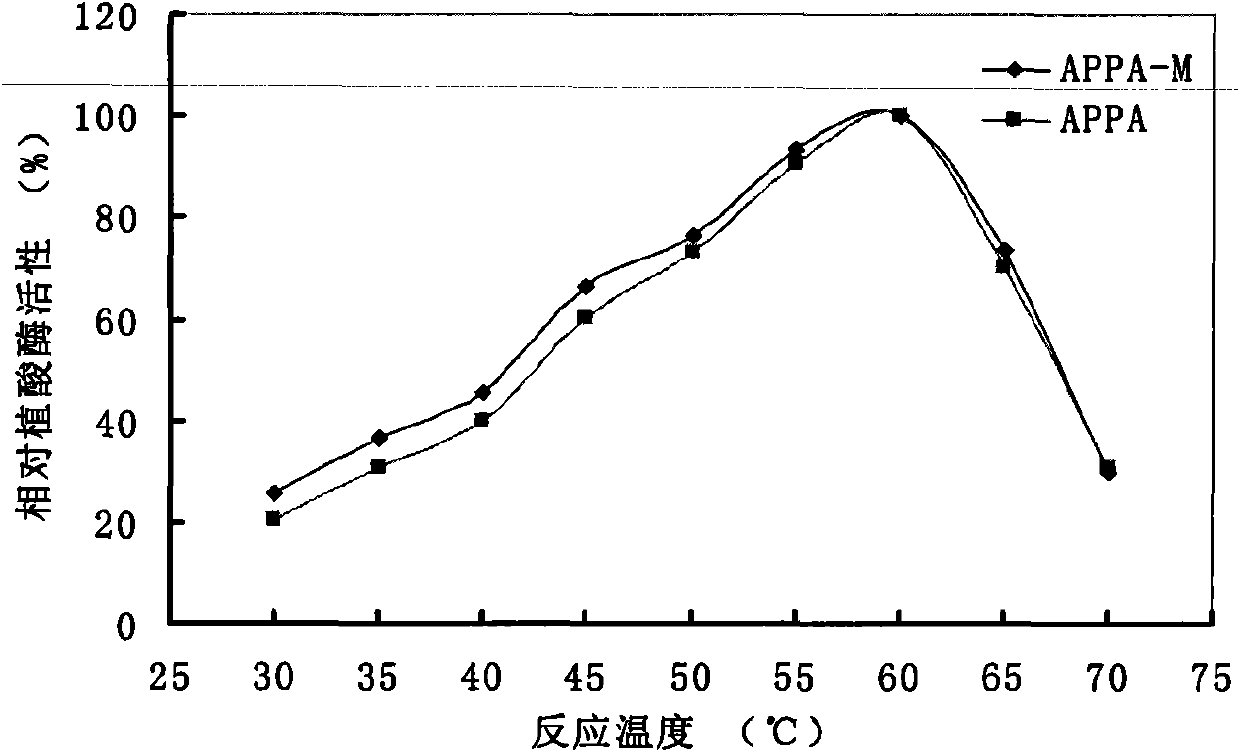
Optimized and improved escherichia coli phytase APPA-M with enhanced catalytic activity in acidic range, and gene and application of optimized and improved escherichia coli phytase APPA-M
A technology of Escherichia coli and phytase, applied in the field of genetic engineering, can solve the problems of waste of phosphorus source, environment, increase of feed cost, etc., and achieve the effect of huge application potential and activity improvement.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1, the synthesis of Escherichia coli phytase gene appa
[0050] The N-terminal signal peptide sequence was removed from the phytase gene appa (Dassa, 1990) of the Escherichia coli strain, and the amino acid sequence was not changed according to the codon preference of Pichia pastoris (Zhao Xiang, 2000). sequence modification. Avoid sites in the form of GT...AG during transformation, and try to avoid the appearance of AT-rich (e.g. ATTTA, AATAA, AATTAA, etc.) sequences, which are related to the stability of mRNA. Send the modified and designed gene sequence to Nanjing Jinruisi Company for whole gene synthesis.
Embodiment 2
[0051] Embodiment 2, construction of recombinant expression vector pET-22b(+)-appa-m
[0052] According to the sequence design of the synthetic gene, the 5' end of the PCR primer contains the Nco I endonuclease site, and the 3' end contains the EcoR I endonuclease site. The primer sequences are as follows:
[0053] 5' end primer pET-appa-F: GCAC CCATGG GACAGAGTGAGCCTGAGTTGAAACTG
[0054] 3' end primer pET-appa-R: GCAC GAATTC TTACAAGGAACAAGCTGGGATTCTAG
[0055] Using the synthetic gene as a template, PCR amplification was carried out with the above primers, and the amplified fragment was cloned into the vector pET-22b(+) to obtain the recombinant vector pET-22b(+)-appa-m.
Embodiment 3
[0056] Embodiment 3, gene site-directed saturation mutation
[0057] (1) Determination of the key amino acids in the active site
[0058] From the protein three-dimensional structure database (http: / / www.rcsb.org / pdb / home / home.do), download the three-dimensional structure file of Escherichia coli phytase APPA, and its number in the PDB database is 1DKQ. The PDB structure is an enzyme-substrate complex and contains the substrate IP6 in its structure. Using PDBViewer to analyze the structure, APPA has a total of 410 amino acid residues, and its 15-23 amino acid residues are the conserved sequence shared by acid phytase: RAGVRAPT. The protein has two structural domains: the 134 amino acid residues at the N-terminal and the 152 amino acid residues at the C-terminal form the alpha domain, and the remaining 124 amino acid residues in the middle form the alpha-beta domain. The conserved sequence and the active center are located in two between domains. Through analysis, 22 key ami...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com