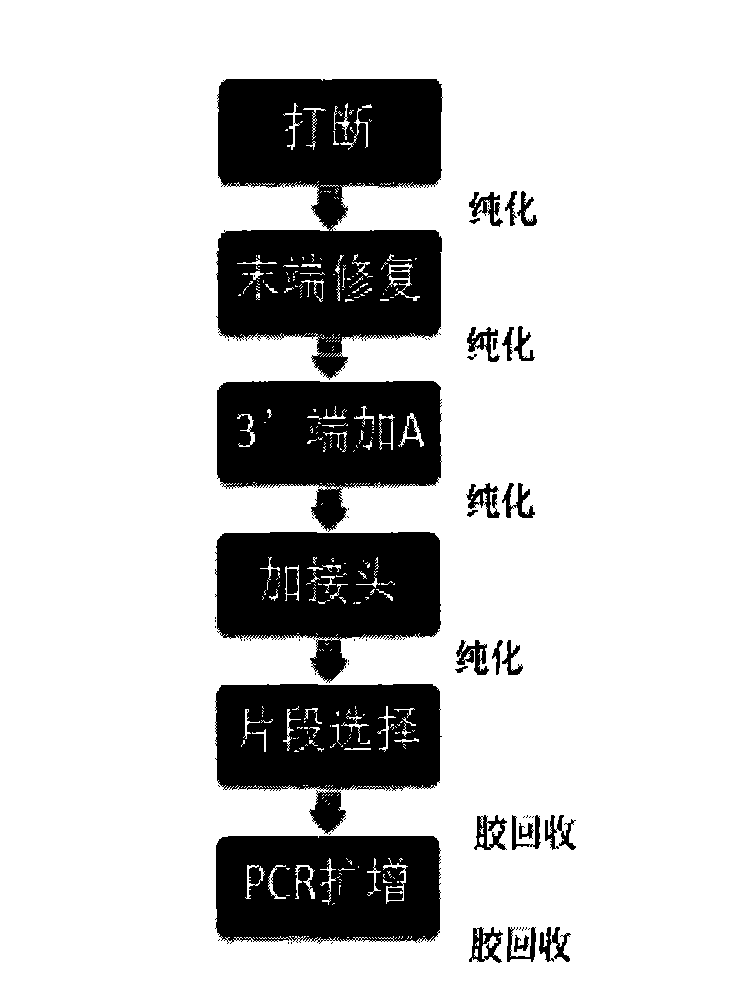
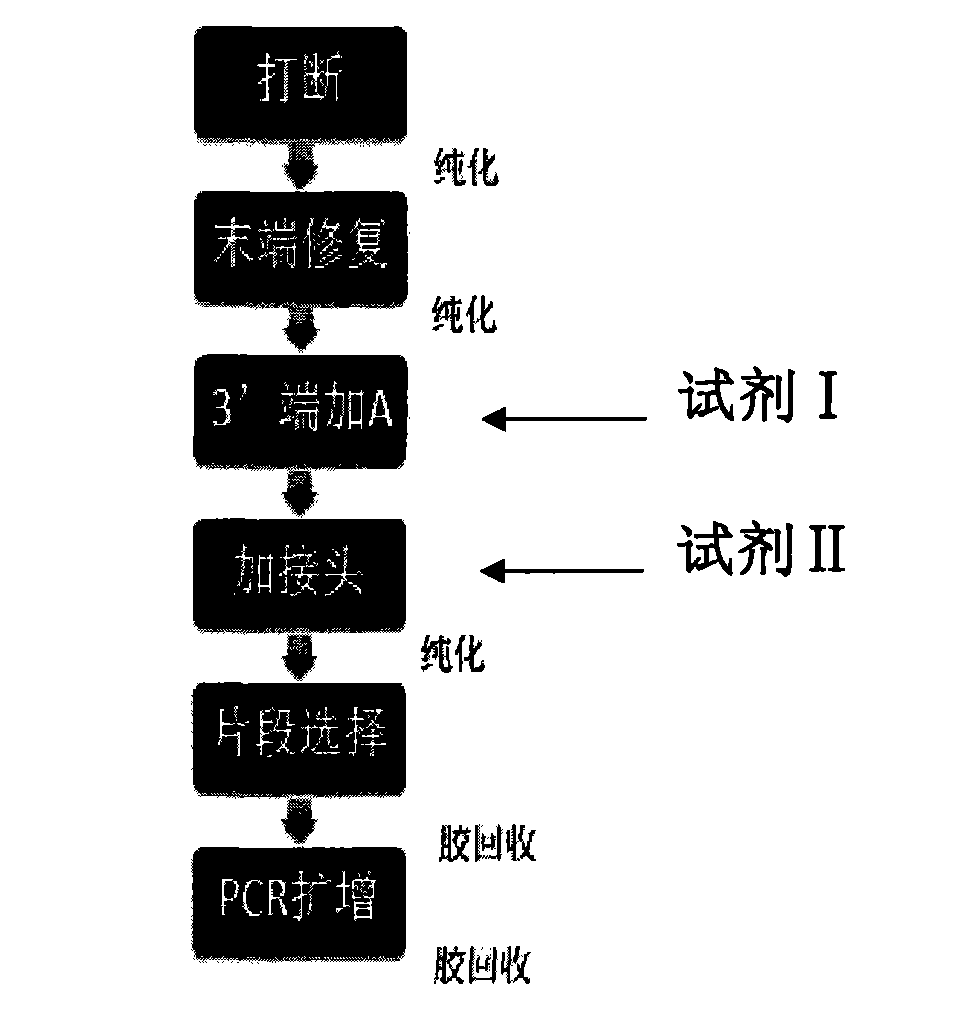
Novel library construction method based on illumina sequencing platform
A sequencing platform and library construction technology, applied in the direction of library, chemical library, nucleotide library, etc., can solve the problems of DNA library sample loss, cumbersome operation process, increased cost, etc., to improve labor efficiency, reduce purification process, improve The effect on success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] 1.1 Reagents
[0077] 10×T4PNK buffer(Enzymatics), dNTP(Enzymatics), T4 DNA polymerase(Enzymatics), T4PNK(Enzymatics), Klenow Fragment(3'to 5'exo)(Enzymatics), Klenow Fragment(Enzymatics), 10Xblue buffer(Enzymatics) , DNA Ligase Buffer 10× (Enzymatics), Phusion DNA polymerase (NEB).
[0078] Reagent I pH is 7.9, and solvent is water, and solute is following final concentration material: 50mM sodium chloride solution (Sinopharm Chemical Reagent Co., Ltd.), 100mM Tris-HCl solution (Sinopharm Chemical Reagent Co., Ltd.), 100mM MgCl 2 (Sinopharm Chemical Reagent Co., Ltd.), 10 mM dithiothreitol (Sigma), 5 mM dATP (Enzymatics), 20 U / ml Klenow Fragment (3′-5′exo).
[0079] The pH of reagent II is 7.6, the solvent is water, and the solute is the following final concentration substances: 72U / ml T4DNA polymerase, 10mM ATP, 500mM Tris-HCl solution, 100mMMgCl 2 , 50mM dithiothreitol;
[0080] 1.2 Experimental steps
[0081] 1.2.1 Take two 50ng / μl Fosmid samples (Shenzhen Huada...
Embodiment 2
[0114] 2.1 Reagents
[0115] 10×blue buffer (Enzymatics), pUC18DNA (Takara).
[0116] Reagent I is the same as that described in Item 1.1 of Example 1.
[0117] Reagent II pH is 7.6, the solvent is water, the solute is T4DNA ligase (Enzymatics), ATP (NEB), 100mM buffered saline solution, 100mM MgCl 2 (Sinopharm Chemical Reagent Co., Ltd.), 50 mM dithiothreitol (Sigma).
[0118] 2.2 Experimental steps
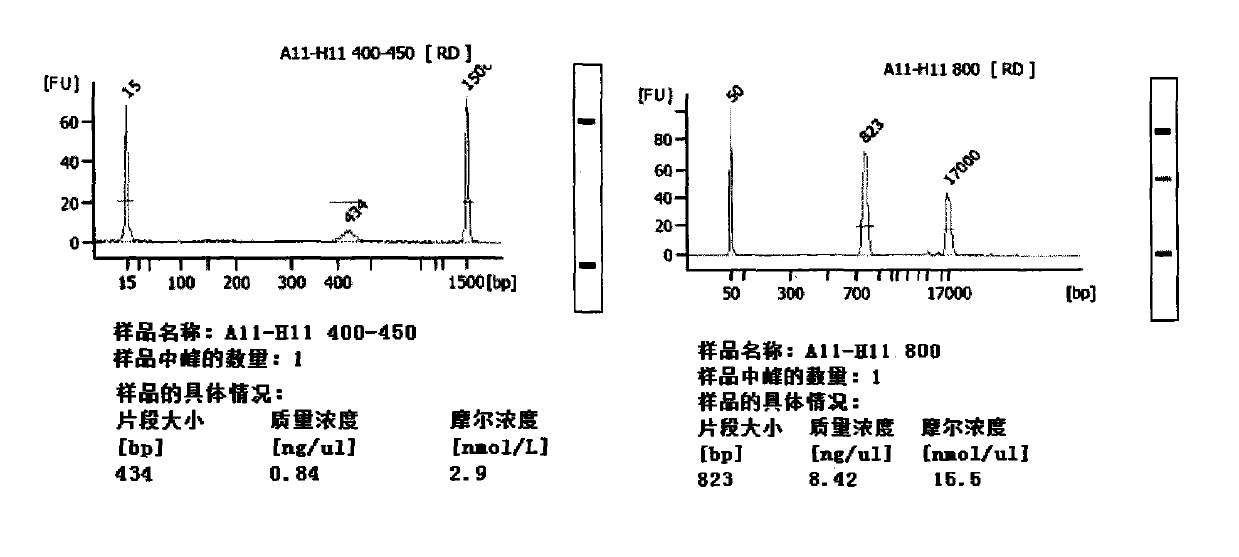
[0119] 2.2.1 Add "A", draw two copies of PCR products (200bp) amplified with pUC18DNA as a template (Shenzhen Huada Gene Research Institute) and place them in EP tubes, name them X and Y respectively, and configure the reaction according to the following table system.
[0120]
[0121] Reaction temperature: 37° C., 30 min. Sample Y was purified by a DNA product purification kit (QIAquick PCR Purification Kit from QIAGEN Company) and dissolved in 16.4 μl of EB eluent (column purification will lose 2 μl of liquid volume).
[0122] 2.2.5 Add "Adapter",
[0123]
[0124]...
Embodiment 3
[0131] 3.1 Reagents
[0132] T4 DNA ligase (Rapid, L603-HC-L) (Enzymatics).
[0133] Reagent I is the same as that described in Item 1.1 of Example 1.
[0134] Reagent II pH 7.6, solvent is water, solute is T4DNA ligase (Takara), 10mM ATP (NEB), 500mM Tris-HCl solution, 100mM MgCl 2 (Sinopharm Chemical Reagent Co., Ltd.), 50 mM dithiothreitol (Sigma).
[0135] 3.2 Experimental steps [Agilent Technologies.Sureselect target enrichment system for illumina paired-end sequencing library.G3360-90020]
[0136] 3.2.1 Take two 50ng / μl Fosmid samples (Shenzhen Huada Gene Research Institute) and place 40μl each in Covaris sample tubes, named X and Y respectively, and use the E210 model breaker produced by Covaris to break the samples. The interrupt parameters are as follows:
[0137]
[0138] 3.2.2 For details on the end repair operation, refer to the Agilent SureSelect platform operation manual G3360-90020.
[0139] 3.2.2 Add "A", and process samples X and Y according to the rea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com