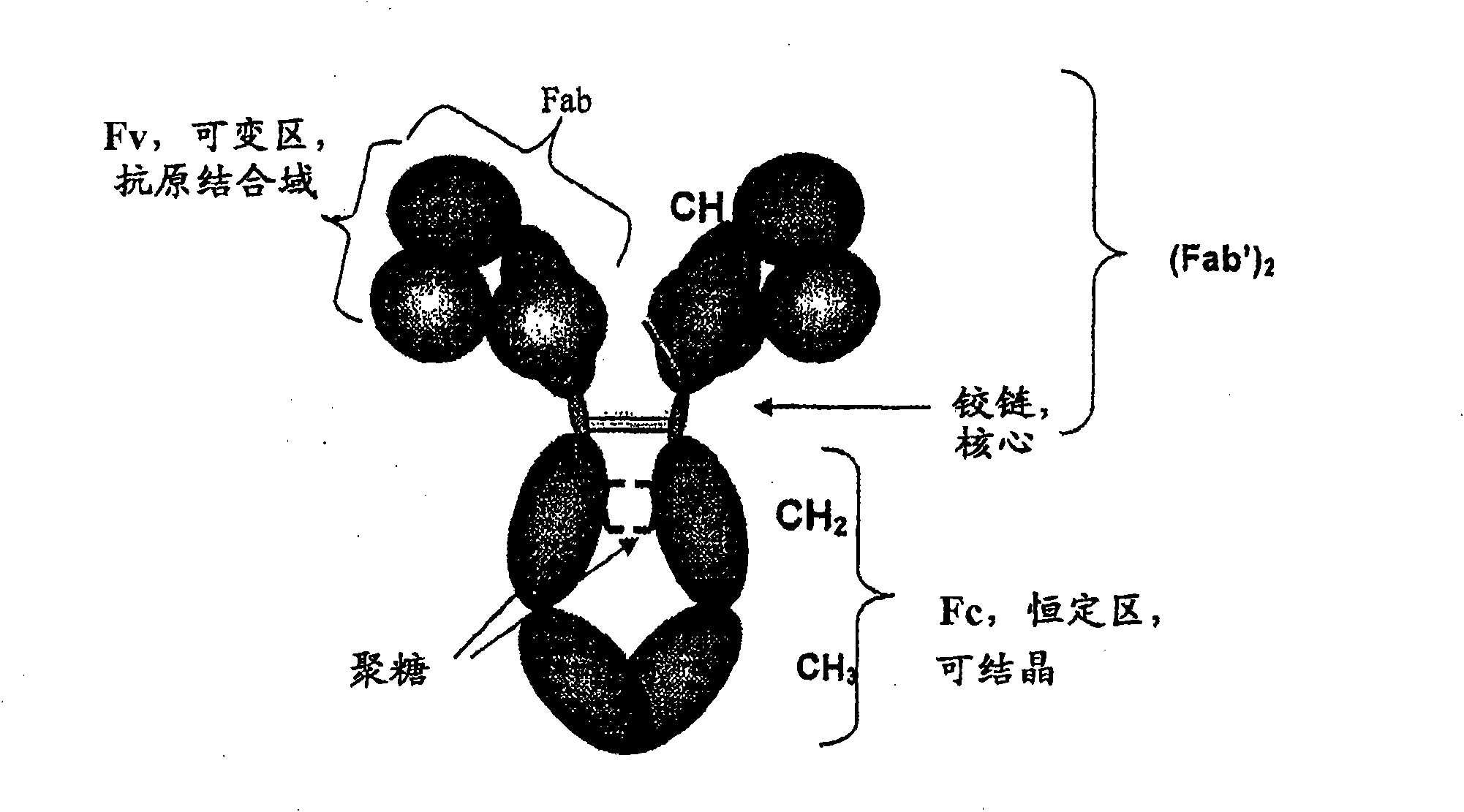
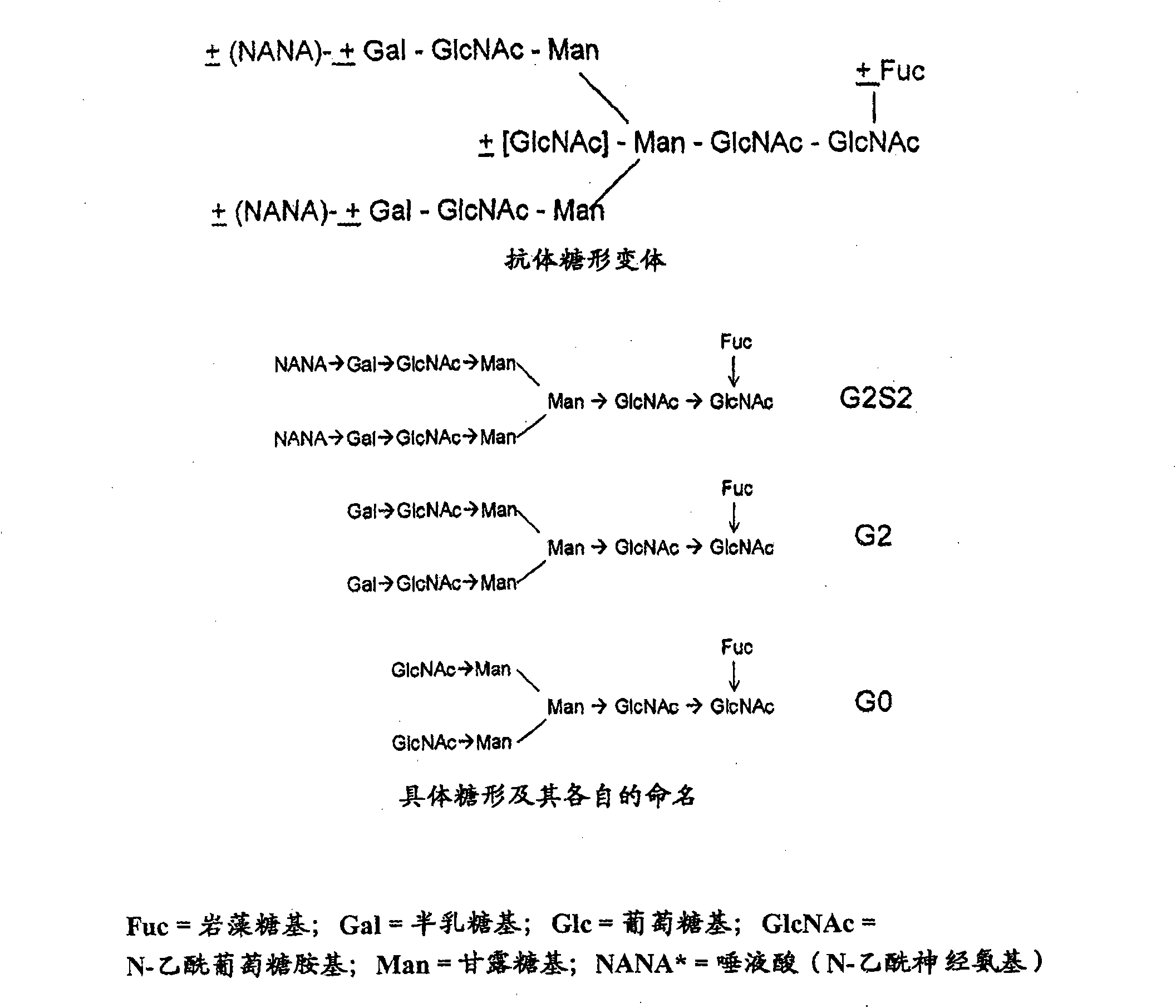
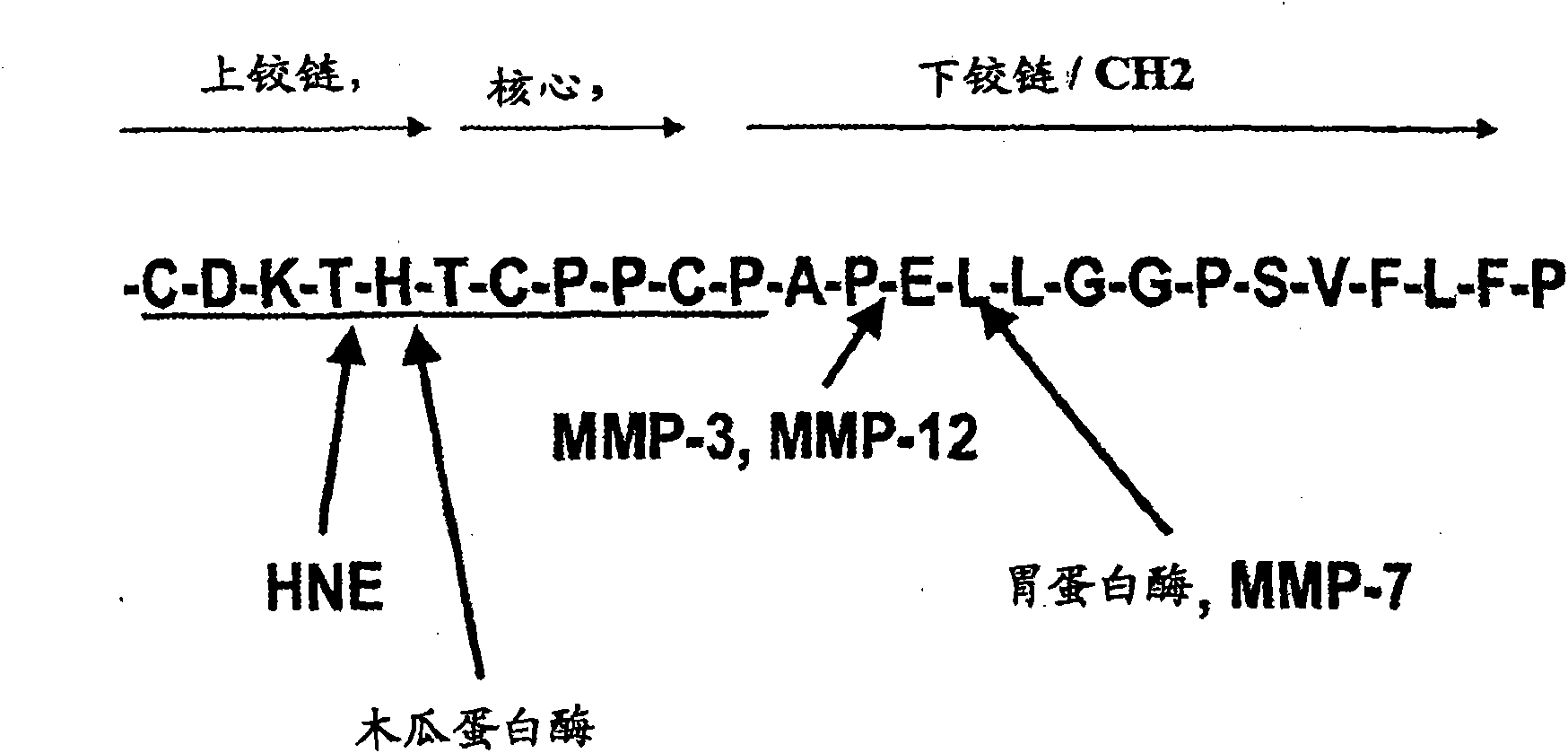
Proteolysis resistant antibody preparations
A protease, elastase technology, applied in the direction of antibodies, anti-inflammatory agents, immunoglobulins, etc., can solve the problem of the existence and composition of unproven glycans
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1: Isolation of FC domain from IgG
[0085] Papain was obtained from Sigma and PNGase F (peptide N-glycosidase F) was obtained from New England Biolabs. Sinapinic acid was obtained from Fluka. MALDI-TOF-MS analysis was performed with a Voyager DE Biospectrometry workstation (Applied BioSystems, Foster City, CA).
[0086] Antibody samples were deglycosylated by treatment with PNGase F in 20 mM Tris-HCl buffer (pH 7.0). Deglycosylated antibody samples were purified on a protein A column (HiTrapA protein cartridges were obtained from Amersham Biosciences) and analyzed for purity by MALDI-TOF-MS.
[0087] Each antibody sample (~1 mg / ml, samples before and after deglycosylation) was treated with papain (1:50, w / w) in 20 mM Tris-HCl buffer (pH 7.0) containing 2 mM L-cysteine For treatment, aliquots were taken at regular time intervals (0, 15, 30, 60, 90 minutes, then 2, 3, 4, 5, 6, 8 and 24 hours). Each aliquot (about 2 μl) was immediately mixed with 2 μl of matri...
Embodiment 2
[0091] Example 2: Papain Digestion of Homogeneous Glycoforms
[0092] To evaluate the parameters of papain digestion of antibody preparations that are substantially homogeneous in their glycosylation pattern, antibody samples were enzymatically modified as described below to generate such preparations for testing.
[0093] To enzymatically galactosylate purified antibody samples, bovine β-1,4-galactosyltransferase (β1,4GT) obtained from Sigma Chemical Co. (St. Louis, MO) and UDP- Gal was added to the antibody sample. Recombinant rat liver α-2,3-sialyltransferase (α2,3ST), recombinant α-1,3-galactosyltransferase (α1,3GT) and CMP-Sia were obtained from Calbiochem (San Diego, CA) . PNGase F was obtained from New England Biolabs (Beverly, MA) or from Prozyme (San Leandro, CA) or from Selectin BioSciences (Pleasant Hill, CA). Diplococcus pneumoniae β-galactosidase and β-glucosaminidase were obtained from ProZyme or from Selectin BioSciences. Bovine kidney β-galactosidase and al...
Embodiment 3
[0101] Example 3: Production of Antibody Fragments Using Matrix Metalloproteinase-3
[0102] Metalloproteases (MMPs) were purified from supernatants of cell clones expressing recombinant human MMPs. The enzyme was activated by treatment with 1 mM 4-aminophenylmercuric acetate (APMA; Sigma) at 37° C. for 1 hour or by treatment with chymotrypsin. Activated enzymes were stored at -70°C. Mix immunoglobulin preparation (0.5-1.0mg / ml) with digestion buffer (250mM Tris-HCl, pH 7.4, containing 1.5M NaCl, 50mM CaCl 2, containing 15-60 μg / ml activated MMP) were incubated at 37°C for 0-24 hours. Aliquots were extracted at 0, 15, 30, 45, 60 and 120 minutes and then at 3, 4, 5, 6, 8, 12 and 24 hours. An aliquot (about 2 μl) was mixed with matrix solution (about 2 μl), and 2 μl of the resulting mixture was applied to the MALDI-TOF-MS target plate, and then MALDI-TOF-MS was performed with a Voyager DE spectrometer. MS analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com