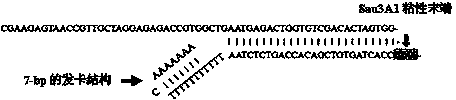Method for detecting insertion flanking sequence and copying quantity of Tgf2 transposon in goldfish genome
A technology of flanking sequences and transposons, which is applied to the detection of Tgf2 transposon insertion flanking sequences and copy numbers in the goldfish genome, achieving high efficiency and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Extraction of fish genomic DNA with Tgf2 transposon insertion
[0041] Due to the insertion of Tgf2 transposon in the goldfish genome, take about 100-200 mg of caudal fin of 95% alcohol or live goldfish, cut it into pieces, and dry at 55°C for 5-6 minutes; add 410 μL of STE lysate (pH=8.10), 80 μL SDS (10%, pH=8.14) and 10 μL of proteinase K (20 μg / ml); turn over and shake well, and keep warm in a 56°C water bath for 14-15 hours; add 340 μL to 400 μL of saturated sodium chloride solution and mix well, shake gently 3min; add 340μL to 400μL of chloroform, shake well; centrifuge at 12000rpm for 20min at 4°C, transfer the supernatant to another clean centrifuge tube; add 450μL to 500μL of isopropanol (-20°C), and centrifuge at 12000rpm for 20min (4 ℃); pour off the supernatant, save the precipitate, add 800μL of 75% ethanol to wash; centrifuge at 12000rpm for 5min to 15min (4℃); to remove ethanol, dry the precipitate, add 100μL to 200μL of double distilled water...
Embodiment 2
[0043] Synthesis of embodiment 2 Splinkerette linker
[0044] Entrusted Shanghai Shenggong Company to synthesize a long oligonucleotide sequence (SEQ ID No: 1) and a short oligonucleotide sequence (SEQ ID No: 2) required for the splinkerette adapter; The short sequence was dissolved in sterile double-distilled water to 50 μm / μl, and 50 μl of oligonucleotide long sequence and short sequence were mixed in equal volumes, incubated at 95°C for 5 minutes, and then gradually lowered to 4°C at 1°C / 15s on the PCR instrument , The long sequence and the short sequence can form a splinkerette joint during the annealing process, and store at -20°C. The synthetic splinkerette linker has the "GATC" cohesive end of restriction endonuclease Sau3A1 on one side, and a 7-bp hairpin structure formed by complementary AT base pairing in the short sequence to form a mismatch region on the other side figure 2 ).
Embodiment 3
[0045] Example 3 Digestion of Goldfish Genomic DNA and Ligation of Splinkerette Adapters
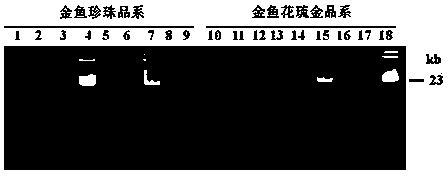
[0046] The goldfish genomic DNA containing the Tgf2 transposon extracted in Example 1 was digested with the restriction endonuclease Sau3A1 in a water bath at 37°C for 12 hours. The digestion reaction was carried out in a 30 μl system containing 10 μl Genomic DNA (2.5 μg), 3μl 10×Sau3A1 Restriction buffer, 4μl Sau3A1 enzyme (10U / μl), 0.3μl Acetylated BSA (10mg / ml), 12.7μl ddH 2 O. The digested products were subjected to 1.2% agarose gel electrophoresis, using TBE as a buffer, and electrophoresis at a voltage of 5V / cm to detect the effect of enzyme digestion ( image 3 ). The Sau3A1 digested DNA product of the complete goldfish genome was placed in a water bath at 65°C for 20 minutes to terminate the activity of the restriction endonuclease, and stored at -20°C for future use. Ligate 4.5 μl of the Sau3A1-digested DNA product and 1 μl of the splinkerette adapter with 4 μl of T4 DNA li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com