Microsatellite DNA molecular markers of lagerstroemia caudate and application
A DNA molecule and microsatellite technology, applied in the field of crape myrtle microsatellite DNA molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 Construction of Lagerstroemia urophylla microsatellite library
[0079] Including the following steps:
[0080] (1) Extract Genomic DNA of Lagerstroemia urophylla (DNA secure Plant Kit, purchased from Tiangen Biotechnology), and digest with endonucleases Rsa I and Xmn I. The enzyme digestion system is: 2.5 μL ligation buffer (NEB), 0.25 μL 100×BSA (NEB), 0.25 μL NaCl (5M), 1 μL Rsa I (NEB), 1 μL Xmn I (NEB), 20 μL DNA (100ng / L), at 37 ° C for 40 hours to obtain a size of 300 ~1000bp DNA fragment;
[0081] (2) Mix 10 μM oligonucleotide SuperSNX24F (5'-GTTTAAGGCCTAGCTAGCAGAATC-3') and terminal phosphorylated SuperSNX24R (5'-pGATTCTGCTAGCTAGGCCTTAAACAAAA-3') in equal volumes, denature at 95°C for 10 minutes, cool slowly to room temperature, and form connector;
[0082] (3) Mix 7 μL of the linker in step (2), 2 μL of T4 DNA ligase (NEB) and 1 μL of 10× ligation buffer, add to the digested product of (1), connect at 22°C for 2 hours, and place in 4 ℃ connected ...
Embodiment 2
[0092] Example 2 Screening and sequencing of positive clones containing Lagerstroemia urophylla microsatellite sequences and design of microsatellite primers for Lagerstroemia urophylla
[0093] Including the following steps:
[0094] (1) Pick 72 white colonies with a toothpick and inoculate them into LB medium containing 50 μg / mL ampicillin, and culture at 37° C. at 150 rpm for 40 hours. Use the bacterial solution as a template to carry out PCR reaction;
[0095] The PCR reaction system is: 2.5 μL 250 μg / mL BSA, 2.5 μL 10×PCR buffer, 1 μL 10 mM primer M13F: 5’-GTAAAACGACGGCCAG-3’, 1 μL 10 mM primer M13R: 5’-CAGGAAACAGCTATGAC-3’, 2 μL 25 mM MgCl 2 , 1.5 μL 2.5mM dNTP, 0.2 μL Taq DNA polymerase, 1.5 μL bacterial solution, 12.8 μL dH 2 O; Reaction program: 95°C for 3min; 95°C for 20sec, 50°C for 20sec, 72°C for 1.5min, 35 cycle reactions; 15°C for heat preservation;
[0096] (2) Detect the reaction product with 1.5% agarose gel electrophoresis, select the bacteria solution with...
Embodiment 3
[0098] Example 3 Using microsatellite DNA molecular markers of Lagerstroemia urophylla to analyze genetic differences among different individuals of Lagerstroemia urophylla
[0099] The specific analysis method is:
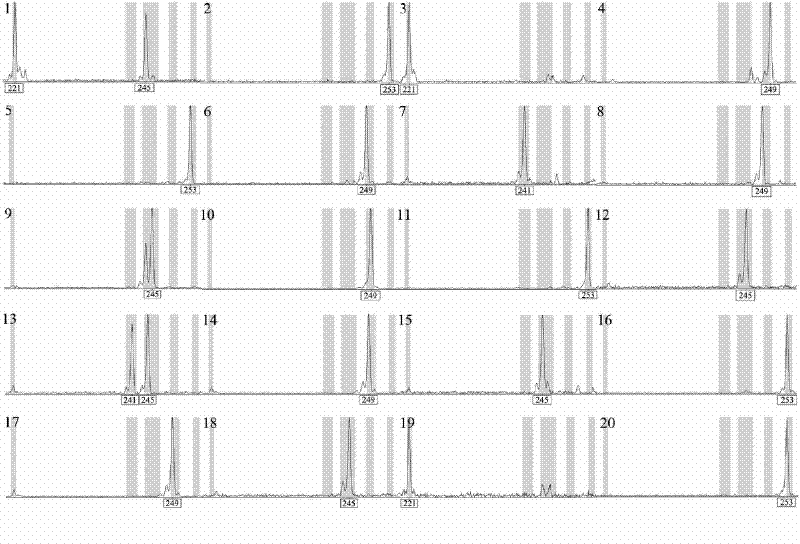
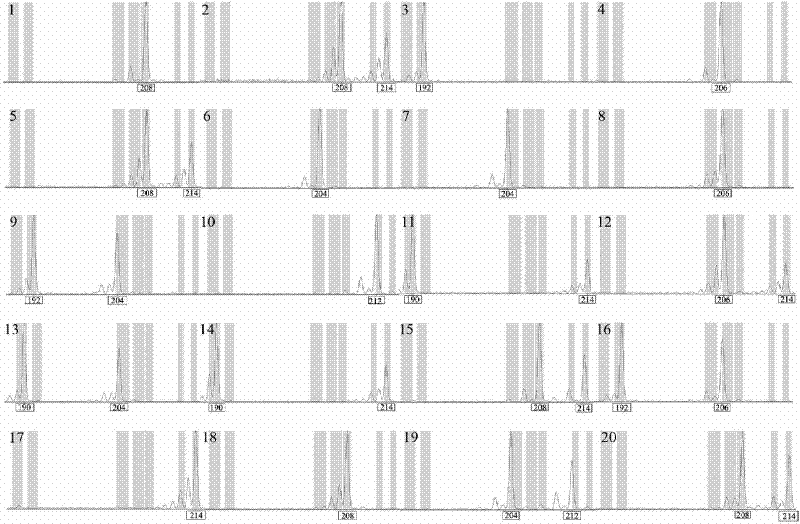
[0100] (1) Carry out the 6' end of the forward primer of the specific primer of I1G, I2B, I2G, I2H, I3G, I9H, I10H, I12H, II1C, II2C, II3A, II3H, II6A, II8A, II12A sites respectively - FAM or JOE mark;
[0101] (2) Genomic DNA of different individual plants of Lagerstroemia urophylla was extracted (DNA secure Plant Kit, purchased from Tiangen Biology);
[0102] (3) Use the specific primers of I1G, I2B, I2G, I2H, I3G, I9H, I10H, I12H, II1C, II2C, II3A, II3H, II6A, II8A, II12A sites to amplify 20 individuals of Lagerstroemia urophylla Genomic DNA. The 10 μL reaction system included 10ng genomic DNA, 1×PCR buffer (10mM Tris-HCl, 50mM KCl, pH 8.3), dNTPs (250μM each), MgCl 2 (1.5mM), forward and reverse primers (0.5μM each), Taq enzyme (0.5U), the reaction program...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com