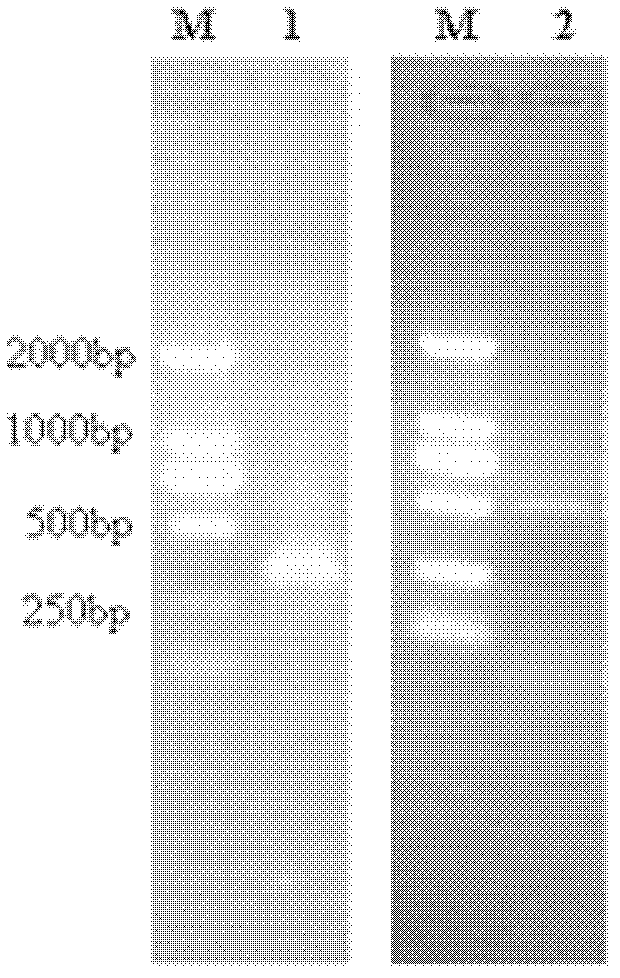Method for preparing human serum albumin-human parathyroid hormone
A technology for parathyroid hormone and human serum albumin, which is used in peptide/protein components, chemical instruments and methods, pharmaceutical formulations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Amplification of the homology arm at the 5' end of YPS1 and the homology arm at the 3' end
[0022] Obtain the 5' end homology arm fragment and the 3' end homology arm fragment of YPS1 sequence from yeast genome by PCR method, used primer YPS1_N up (SEQ ID NO.1), YPS1_N dn (SEQ ID NO.2) and YPS1_C up (SEQ ID NO.3) and YPS1_C dn (SEQ ID NO.4) were synthesized with an oligonucleotide synthesizer. The upstream and downstream primers for the amplification of the homology arm at the 5' end are respectively introduced into the PstI and SalI sites and the protective bases, and the upstream and downstream primers for the amplification of the 3' homology arms are respectively introduced into the BglII and PstI sites and the protective bases base, the endonuclease recognition sequence is underlined.
[0023] YPS1_N up: 5'-aagc ctgcag agctccattg cgccaacccc-3'
[0024] YPS1_N dn: 5'-cggc gtcgac aatctggctg agcggaaagt ttga-3'
[0025] YPS1_C up: 5'-ccc agatct c a...
Embodiment 2
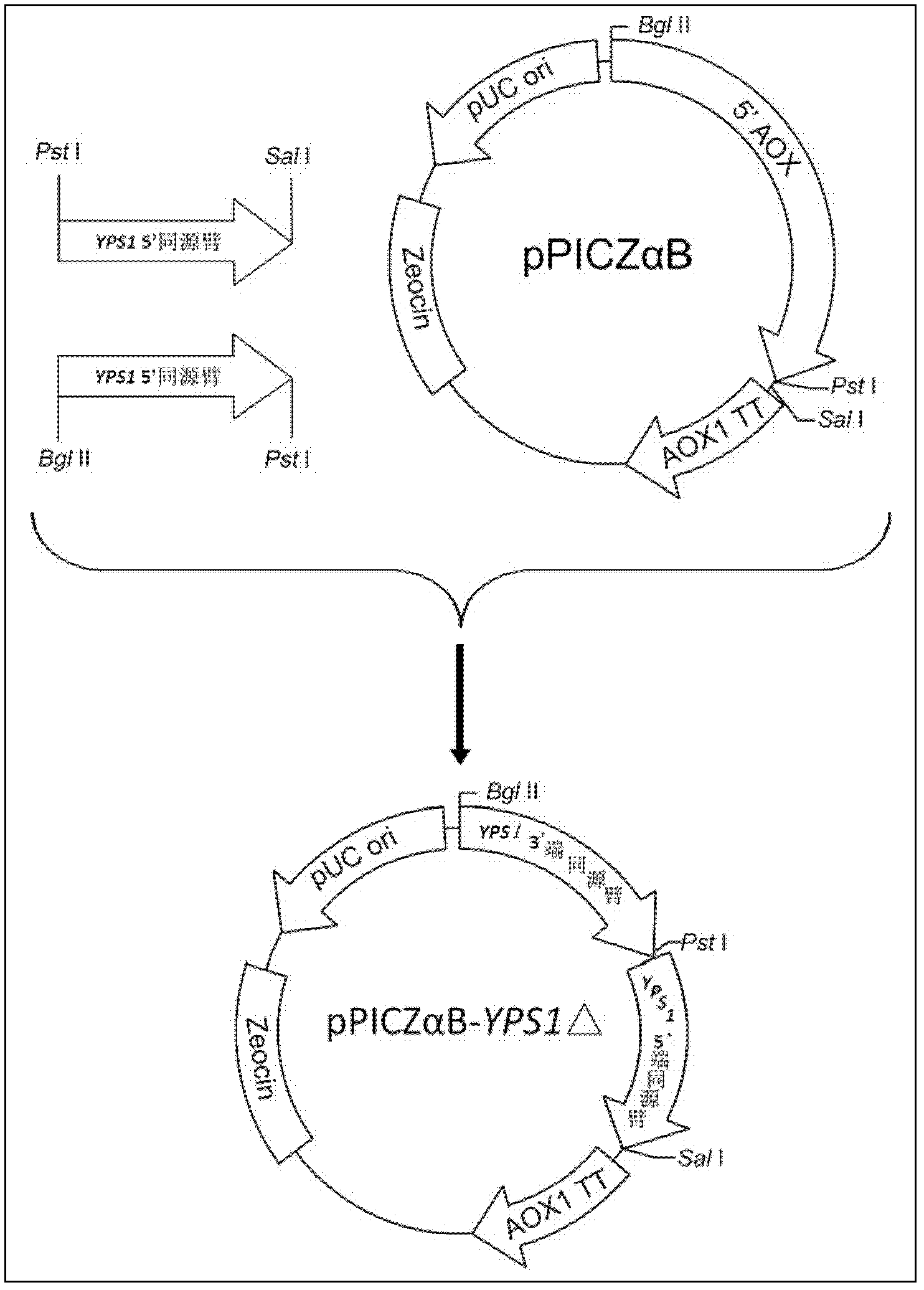
[0028] Example 2: Construction of recombinant vector pPICZαB-YPS1Δ
[0029] After electrophoresis purification of the 3' homology arm PCR product obtained in Example 1, the target band was recovered and purified with a DNA fragment recovery kit. The purified 3' homology arm PCR product was digested with BglII / PstI and recovered by electrophoresis; the plasmid pPICZαB was also digested with BglII / PstI, and the linearized fragment was recovered by electrophoresis. The digested 3' homology arm and the vector pPICZαB were mixed in an appropriate ratio, and ligated with T4 ligase overnight in a water bath at 16°C to form pPICZαB-C. After transformation into DH5α, spread on LB agar plates containing 25 μg / mL Zeocin, and culture overnight at 37°C. Select several clones on the plate and inoculate them into 5 mL LB liquid medium containing 25 μg / mL Zeocin, and culture overnight at 37°C. The positive clone pPICZαB-C / DH5α was obtained by PCR verification of YPS1_C up / YPS1_C dn with pri...
Embodiment 3
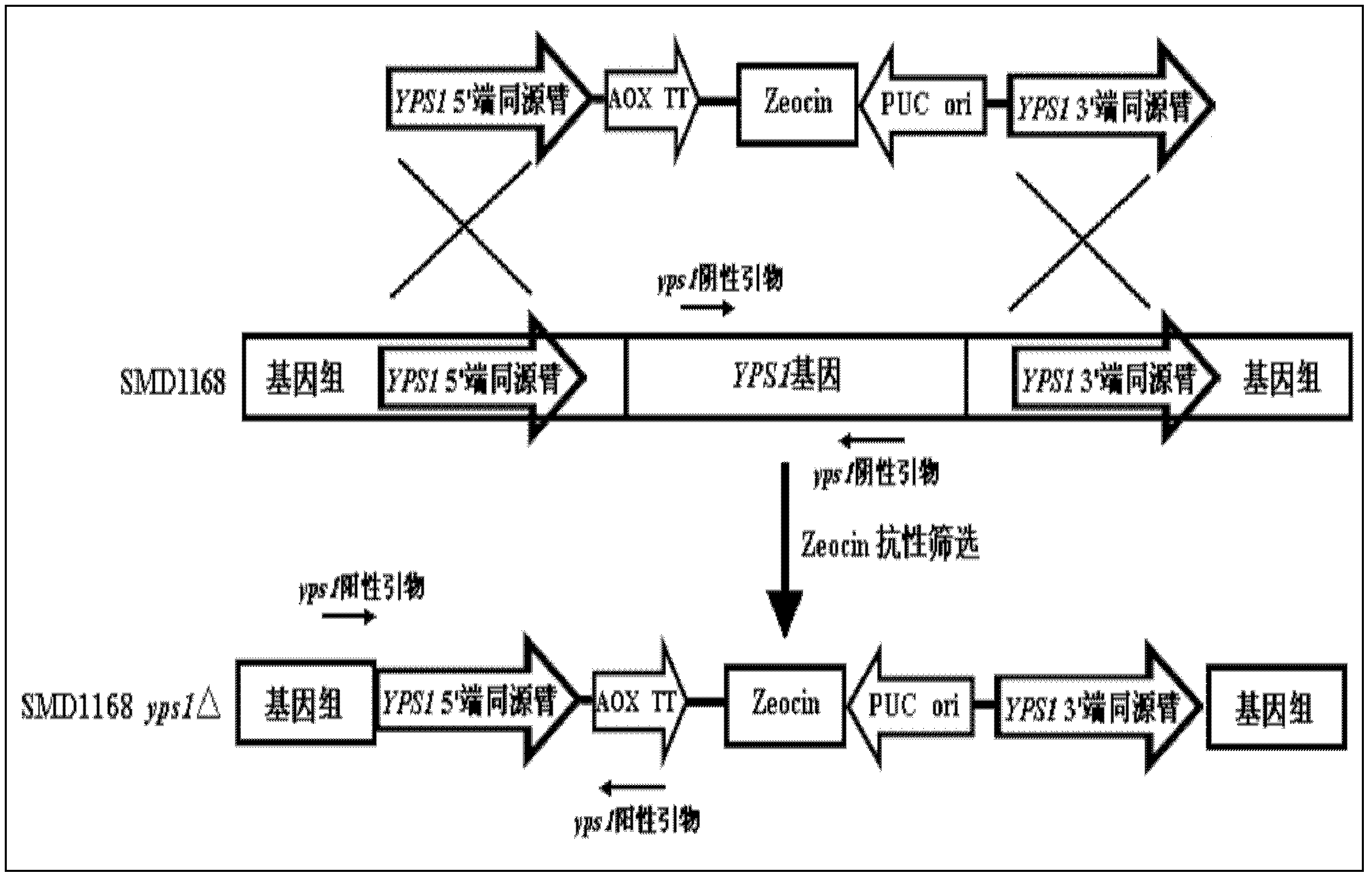
[0031] Example 3: Homologous recombination method to knock out the YPS1 gene in the host SMD1168
[0032] The Pichia host SMD1168 is a protease A-deficient host. Therefore, on the basis of SMD1168, the aspartate hydrolase 1 gene can be further knocked out to obtain a Pichia host with a double knockout of protease A and yapsin 1.
[0033] After linearizing pPICZαB-YPS1Δ with PstI, it was transformed into Pichia pastoris SMD1168 by electroporation (the yeast expression kit was provided by Invitrogen Corp. / mL Zeocin and 1M sorbitol on YPD agar plate (1% yeast extract, 2% peptone, 2% glucose, 1M sorbitol, 2% agarose), culture at 30°C for 3-5 days. Pick several clones on the plate and inoculate them in 5 mL of YPD liquid medium containing 50 μg / mL Zeocin, and culture overnight at 30°C. PCR verification of positive clones, see image 3 , Figure 4 : Design a pair of negative primers (yps1 negative up (SEQ ID NO.5), yps1 negative dn (SEQ ID NO.6)), a pair of positive primers (yps...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com