Single-stranded deoxyribonucleic acid (ssDNA) aptamer targeting human highly metastatic hepatoma carcinoma cell and application thereof
A technology of liver cancer cells and aptamers, which can be used in recombinant DNA technology, DNA/RNA fragments, medical preparations with non-active ingredients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
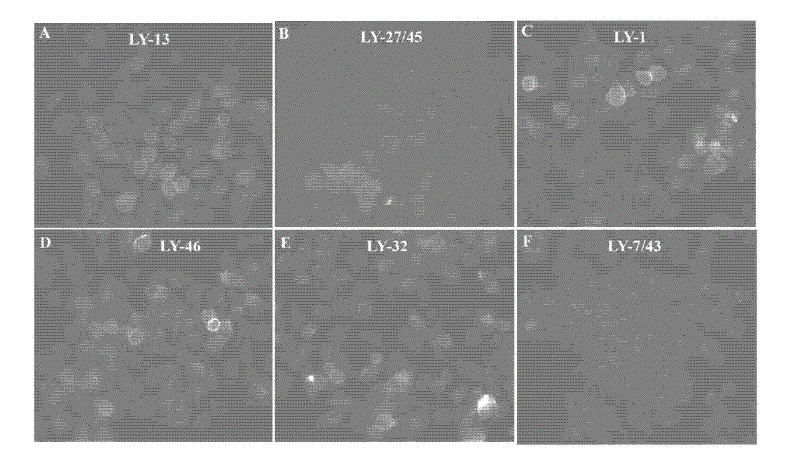
[0082] Example 1. Subtractive cell-SELEX screening of nucleic acid aptamers that specifically bind to highly metastatic liver cancer cells
[0083] 1.1 Pre-screening treatment of high metastatic potential liver cancer cells HCCLM9 (target cells) and low metastatic potential liver cancer cells MHCC97-L (subtractive cells)
[0084] Cells in good growth condition were treated with enzyme-free cell digestion solution, and 5×10 6 Cells were inoculated in a culture dish and cultured overnight. When the fusion rate of the cells reached 95%, they could be used for library screening.
[0085]1.2 The first round of cell selection
[0086] 1.2.1 Select HCCLM9, a monolayer adherent overnight growth cell with a cell fusion rate of 95%, as a screening target, use trypan blue to identify its activity, and use 1×PBS-1mmol / LMgCl 2 -0.1%BSA rinse cells twice;
[0087] 1.2.2 Incubate the random ssDNA library with the target cells on ice for 60 minutes, so that the ssDNA library can fully inte...
Embodiment 2
[0147] Example 2. Recognition of liver cancer cells based on quantum dot-aptamer functionalized biomolecular probes
[0148] 2.1 Animal model making:
[0149] According to the method of this group, the animal model of lung metastasis of human liver cancer was made, and the brief description is as follows:
[0150] 2.1.1 Observe that the MHCC97-L and HCCLM9 cells grow well, and the cell fusion rate reaches 80%. Trypsinize them respectively, put them in 15ml centrifuge tubes, centrifuge at 1000rpm×10min at 4°C, discard the supernatant, and press 5×10 6 / 0.2mL dose was inoculated subcutaneously on the back of nude mice, and subcutaneously formed tumors after 2 weeks, with a diameter of about 0.5cm.
[0151] 2.1.2 Orthotopic implantation: Simultaneously excise the subcutaneous tumors on the back of MHCC97-L and HCCLM9 nude mice, and place them in culture dishes added with 1×PBS respectively. Intrahepatic orthotopic tumor inoculation, the method of MHCC97-L and HCCLM9 is the same...
Embodiment 3
[0202] Example 3. Peripheral blood simulated environment circulating liver cancer cell enrichment test
[0203] 3.1 Red blood cell lysis
[0204] 10 respectively 4 a, 10 2 Put a highly metastatic liver cancer cell HCCLM9 into 1mL peripheral whole blood of a healthy person, add 2 times the volume of PBS, mix well, 1500r / min, 5min, remove the supernatant, add red blood cell lysate NH 4 Cl, incubate at room temperature for 10min, 1500r / min, 5min, remove the supernatant; repeat lysis of residual red blood cells once, 1500r / min, 5min, remove the supernatant, add 10 times the volume of PBS to wash once, remove the supernatant, add the adapter Sub-binding solution 1mL, mix the cell pellet.
[0205] 3.2 Incubation of aptamers and mixed cells
[0206] Add a certain concentration of aptamers to make the final concentration 500pM, incubate on ice for 30min with shaking, wash with ice-cold PBS 3 times / 5min, 1500r / min, 5min, remove the supernatant for the last time, and cell pellet for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com