Erythropoietin mimetic peptide (EMP)-human serum albumin (HSA) fusion protein and preparation method thereof
A technology of human serum albumin and erythropoietin, which is applied in the field of biotechnology and genetic engineering pharmaceuticals, and can solve problems such as difficult expression and preparation, small molecular weight, and poor application effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
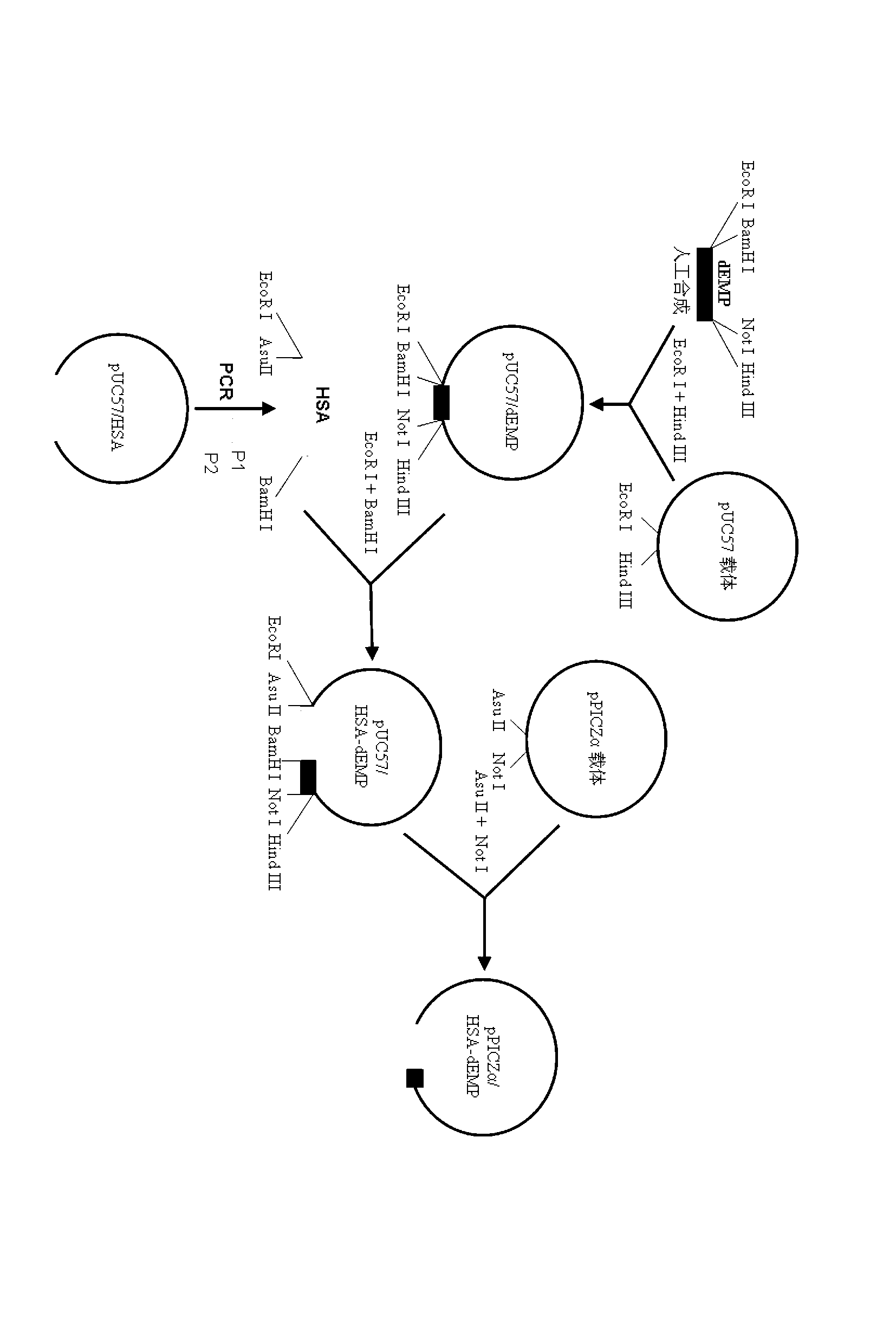
[0078] Example 1: Artificial synthesis of cDNA sequence encoding EMP doublet and connecting peptide
[0079] 1. First derive the corresponding coding nucleotide sequence according to the amino acid sequence of the EMP dimer and the connecting peptide, and optimize the coding sequence according to the Pichia pastoris preferred codons. In order to facilitate the fusion with the HSA gene, the selection will be tight The coding sequence of the two amino acids (Gly, Ser) of HSA is designed as restriction endonuclease BamH I recognition site (gga tcc); at the same time, in order to facilitate cloning, the coding nucleotide sequence is introduced at the 5'end EcoR Recognition site and protective bases are cut with I restriction, introduced at the 3'end Not I and Hind Recognition sites and protective bases for restriction enzyme III, the final nucleotide sequence is as follows (SEQ ID NO: 7):
[0080] agc gaa ttc gga tccggc gga ggt gga tct gga ggc ggt gga tct ggt ggt ggaact tat ...
Embodiment 2
[0083] Example 2: Obtaining cDNA fragments encoding HSA
[0084] 1. Design and synthesize PCR primers for HSA gene
[0085] P1 (SEQ ID NO: 8): 5′-agc gaa ttc ttc gaa acg atg aag tgg gta acc ttt att tcc ctt c -3′
[0086] P2 (SEQ ID NO: 9): 5′-gat gga tcc taa gcc taa ggc agc ttg ac- 3′
[0087] The underlined bases in P1 are restriction endonucleases EcoR I Recognition site sequence, the shaded bases are restriction endonucleases Asu II recognition site sequence, the framed part of the base is the translation initiation codon sequence; the shaded part in P2 is the restriction endonuclease BamH I's recognition site sequence.
[0088] 2. PCR amplification
[0089] Use pUC57 / HSA plasmid DNA as a template (pUC57 / HSA plasmid is preserved in this room, see invention patent 201110420870.0 for the specific construction method), and P1 and P2 are used as upstream and downstream primers for PCR amplification. The reaction conditions are as follows: ①denaturation: 94℃, 5min; ②denaturation...
Embodiment 3
[0090] Example 3: pPICZα / Construction of HSA-dEMP recombinant expression vector
[0091] 1. Cloning and sequencing of the cDNA fragment encoding HSA-dEMP:
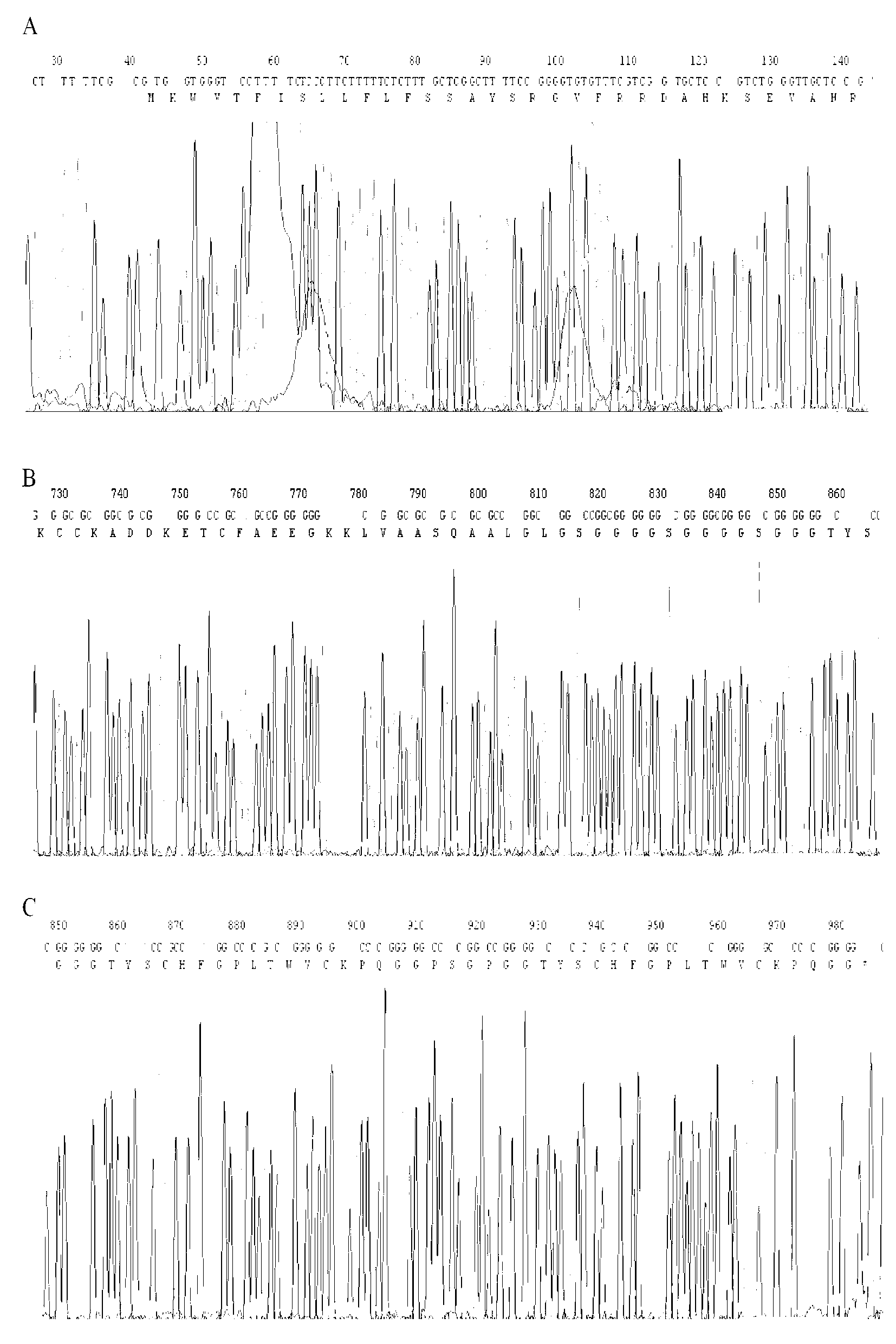
[0092] The pUC57 / dEMP plasmid DNA obtained in Example 1 and the PCR product of HSA obtained in Example 2 were performed simultaneously EcoR I + BamH I double enzyme digestion, the digested DNA fragment is recovered by the gel, and then the T4 DNA ligase is used for ligation reaction. The ligation product is transformed into E. coli DH5α competent cells, coated on an ampicillin resistant LB plate and incubated overnight at 37°C to select positive clones. The obtained clone was sent to Shanghai Shenggong Bioengineering Company for sequencing, and the clone with the correct sequence was named pUC57 / HSA-dEMP.
[0093] 2. Construction of yeast expression vector pPICZα / HSA-dEMP
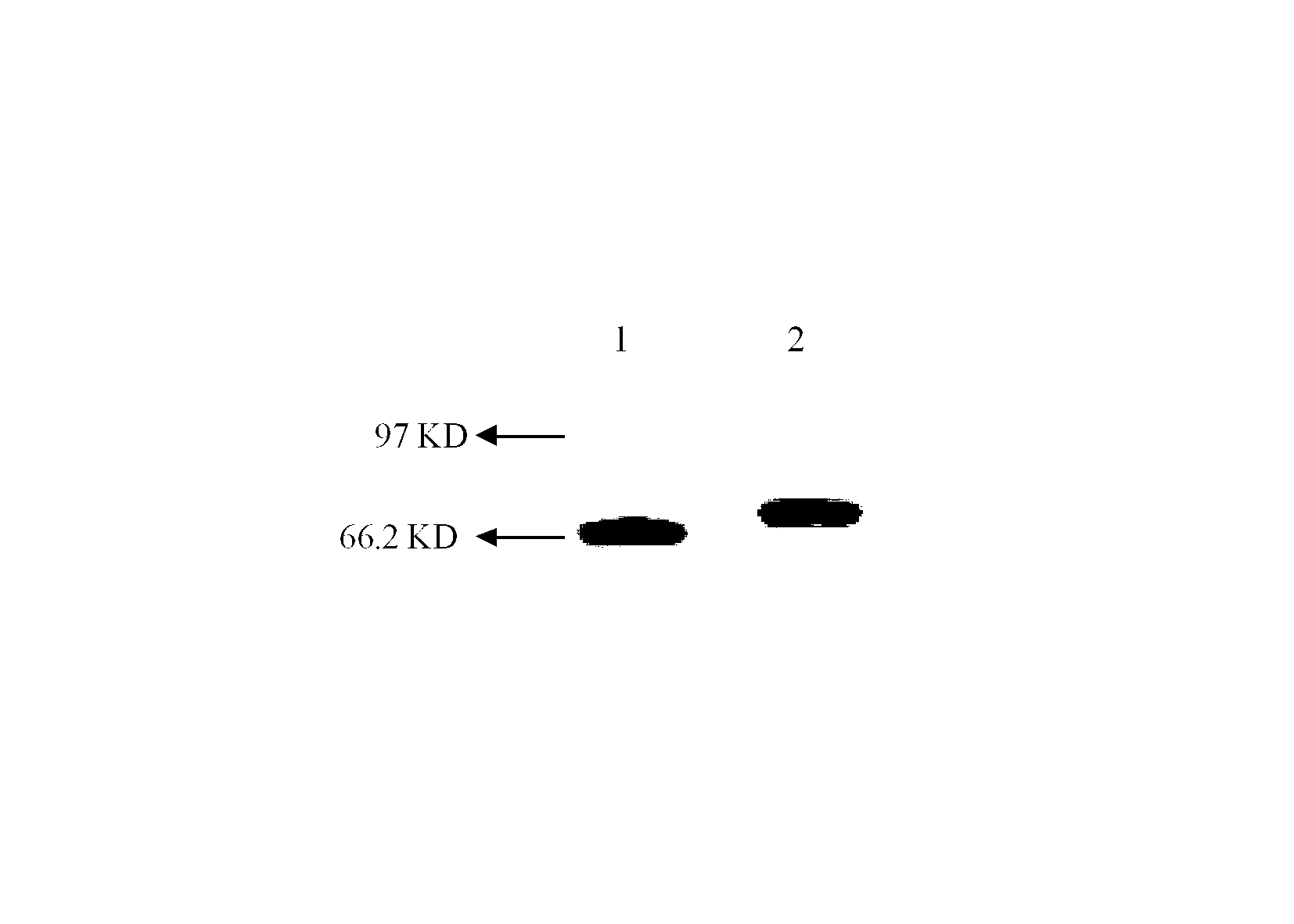
[0094] Extract the pUC57 / HSA-dEMP plasmid DNA sequenced in the previous step, Asu II and Not I double-enzyme digestion of plasmid DNA, and gel reco...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com