Gene chips for detecting multiple pathogenic bacteria in animals cultivated in sea water and uses thereof
A gene chip and mariculture technology, which is applied to the gene chip and application field for detecting various pathogenic bacteria of marine aquaculture animals, can solve the problems of lack of evidence and persuasiveness in the virulence analysis of pathogenic bacteria and so on.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0087] Then follow the following specific steps for chip preparation, sample preparation, chip hybridization, and acquisition and analysis of hybridization results:
[0088] 1. Chip preparation:
[0089] Dissolve the probe modified with the 3'end amino group to a final concentration of 10-100μM, mix it with 30-60% DMSO (dimethyl sulfoxide), and dilute it into a spot sample with a final probe concentration of 10-30μM. The spotter performs the spot system of the chip, and the layout of the probe array on the chip is shown in Table 3. After the probe is spotted and fixed according to the instructions of the aldehyde-based or amino-based substrate fixing process.
[0090] Second, the application of the chip
Embodiment 1
[0091] Example 1: Effect detection of the chip
[0092] (1) Extraction of DNA from samples to be tested:
[0093] Randomly select 8 species of pathogenic bacteria in marine aquaculture animals: Vibrio anguillarum, Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio fischeri, Vibrio splendidu, Aeromonas hydrophila, Aeromonas salmonicida, Edwards tardy Bacteria, and calf thymus DNA are tested for the effect of gene chip.
[0094] Use liquid culture medium to cultivate pathogenic bacteria overnight at an appropriate temperature, use bacterial genomic DNA extraction kit or traditional methods to extract bacterial genomic DNA, and dilute it to 1.0ng / μL as a PCR reaction template.
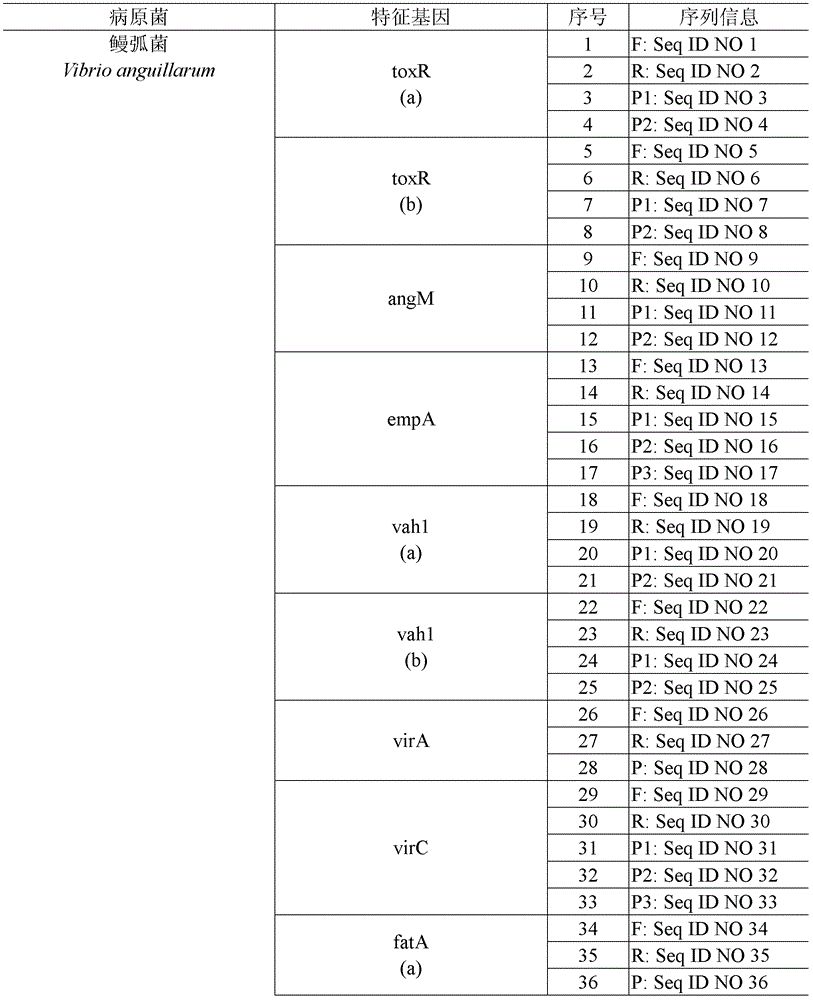
[0095] Randomly select 14 characteristic genes of 8 pathogens and 3 genes of the quality control system for synchronous gene chip testing. The primers and probes corresponding to these 17 genes are as follows:
[0096] Table 5. Primers and probes information corresponding to 14 genes of 8 mariculture animal pat...
Embodiment 2
[0120] Example 2: Application of the chip
[0121] (1) Extraction of DNA from samples to be tested
[0122] The gill tissue of a diseased turbot of a farm in Shandong was taken, half of which was extracted using the marine animal tissue genomic DNA extraction kit produced by Tiangen Biochemical Technology Co., Ltd.; the other half was used for bacterial analysis on 2216E seawater medium Cultivation and single colony screening, retain one of the dominant bacteria, and use the bacterial genomic DNA extraction kit to extract the genomic DNA of the bacteria, which is also used for gene chip detection.
[0123] (2) PCR synchronous amplification of target sequences of characteristic genes and quality control system genes:
[0124] Use all the primers in Table 5 and the 16S rRNA primers in Table 6 to simultaneously amplify the template to be tested, and add a separate primer pair to each PCR tube. The amplification reaction solution includes genomic DNA from the gill tissue of turbot, 1.0μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com