Primer and method for PCR-DHPLC (polymerase chain reaction-denaturing high-performance liquid chromatography) detection for endogenous genes of wheat
A PCR-DHPLC and endogenous gene technology is applied to the PCR-DHPLC detection primers and detection fields of wheat endogenous genes, and achieves the effects of a reliable detection method, good expansion performance, high sensitivity and resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 DNA Extraction
[0031] Sample DNA was extracted by CTAB method, as follows:
[0032] a) Weigh 5 g of the sample, add liquid nitrogen to the mortar and grind until the sample is a powder with a size of about 0.5 mm;
[0033] b) Weigh 300 mg of the ground sample, quickly transfer it to a 2 mL centrifuge tube, add 700 μL of CTAB extract solution preheated at 65 °C, mix well, and put it in a water bath at 65 °C for 30 min;
[0034] c) Add 5 μL RNase (10 mg / mL), and bathe in water at 37°C for 30 minutes;
[0035] d) Add an equal volume of Tris saturated phenol, mix thoroughly, and centrifuge at 12000r / min for 15min;
[0036] e) Take the supernatant, add an equal volume of chloroform / isoamyl alcohol (24:1) to mix, and centrifuge at 12000r / min for 15min;
[0037] f) Take the supernatant, add an equal volume of chloroform / isoamyl alcohol (24:1) to mix, and centrifuge at 12000r / min for 15min;
[0038] g) Add an equal volume of pre-cooled isopropanol, shake gently,...
Embodiment 2
[0041] Example 2 DNA concentration determination
[0042] The concentration and purity of the extracted sample DNA were measured; the absorbance values at 260nm and 280nm were measured by an ultraviolet spectrophotometer, and the purity and concentration of nucleic acid were calculated respectively. The calculation formula is as follows:
[0043] DNA purity = OD260 / OD280
[0044] DNA concentration=50×OD260mg / mL
[0045] The purity ratio of DNA was between 1.7 and 1.9, and the concentration was greater than 10ng / μL.
Embodiment 3
[0046] Embodiment 3 PCR amplification
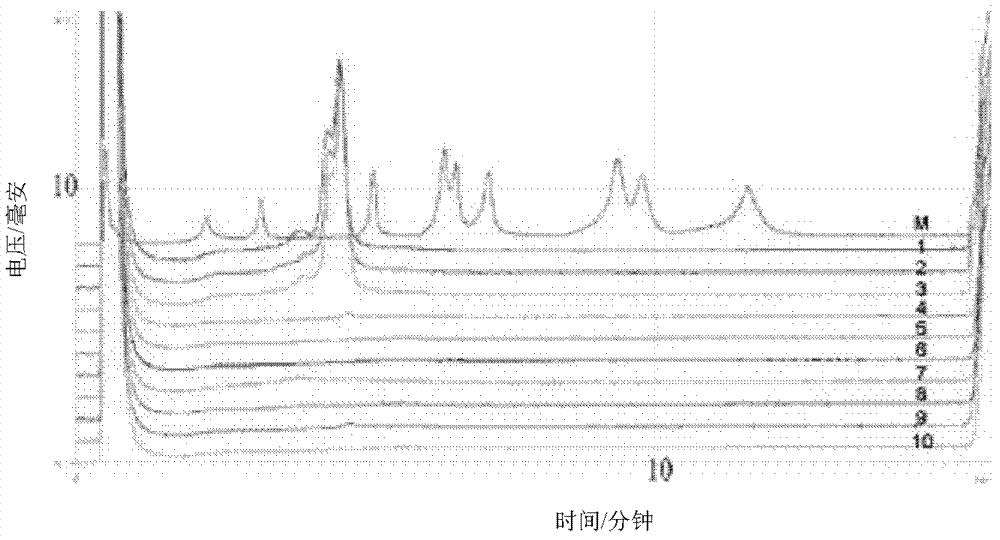
[0047] Design the binding site of the specific detection primer according to the wheat endogenous gene acc1, and add the regulatory sequence at the 5' end of the specific detection primer binding site, and synthesize the detection primer containing the regulatory sequence (Table 1). PCR amplification with upstream / downstream primers, sample settings include: DNA from 3 wheat samples and DNA from non-GM rapeseed, DNA from potato line EH92-527-1, DNA from non-GM rice, DNA from non-GM cotton, DNA from non-GM DNA from corn, DNA from papaya and water blank.
[0048] Table 1 The detection primer binding sites and primers of wheat endogenous gene acc1
[0049]
[0050]
[0051] The total volume of the PCR reaction system is 50 μL, and the components are: multiplex PCR reaction mixture Multiplex PCR Mix (TaKaRa) 25 μL, 10 μmol / L primers 1 μL, DNA 2 μL, 5U / μL Taq enzyme 0.25 μL, with sterilized double distilled water Make up to 50 μL.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com