Nucleic acid aptamer, complementary sequence and detection method for detecting hemolytic streptococcus
A technology of hemolytic streptococcus and nucleic acid aptamer, applied in the field of clinical testing, can solve the problems of expensive reagents, difficult to popularize, etc., and achieve the effects of large detection volume, direct and objective results, and simple operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] 1. Experimental materials
[0034] 1.1 Fluorescence spectrophotometer measurement (Hitachi, Japan F7000); transmission electron microscope (JEM-1230, JEOL, Japan); pH meter (Shanghai REX Instrument Factory)
[0035] 1.2 Culture medium: brain heart infusion broth and blood plate are produced by Guangdong Huankai Microbial Technology Co., Ltd.
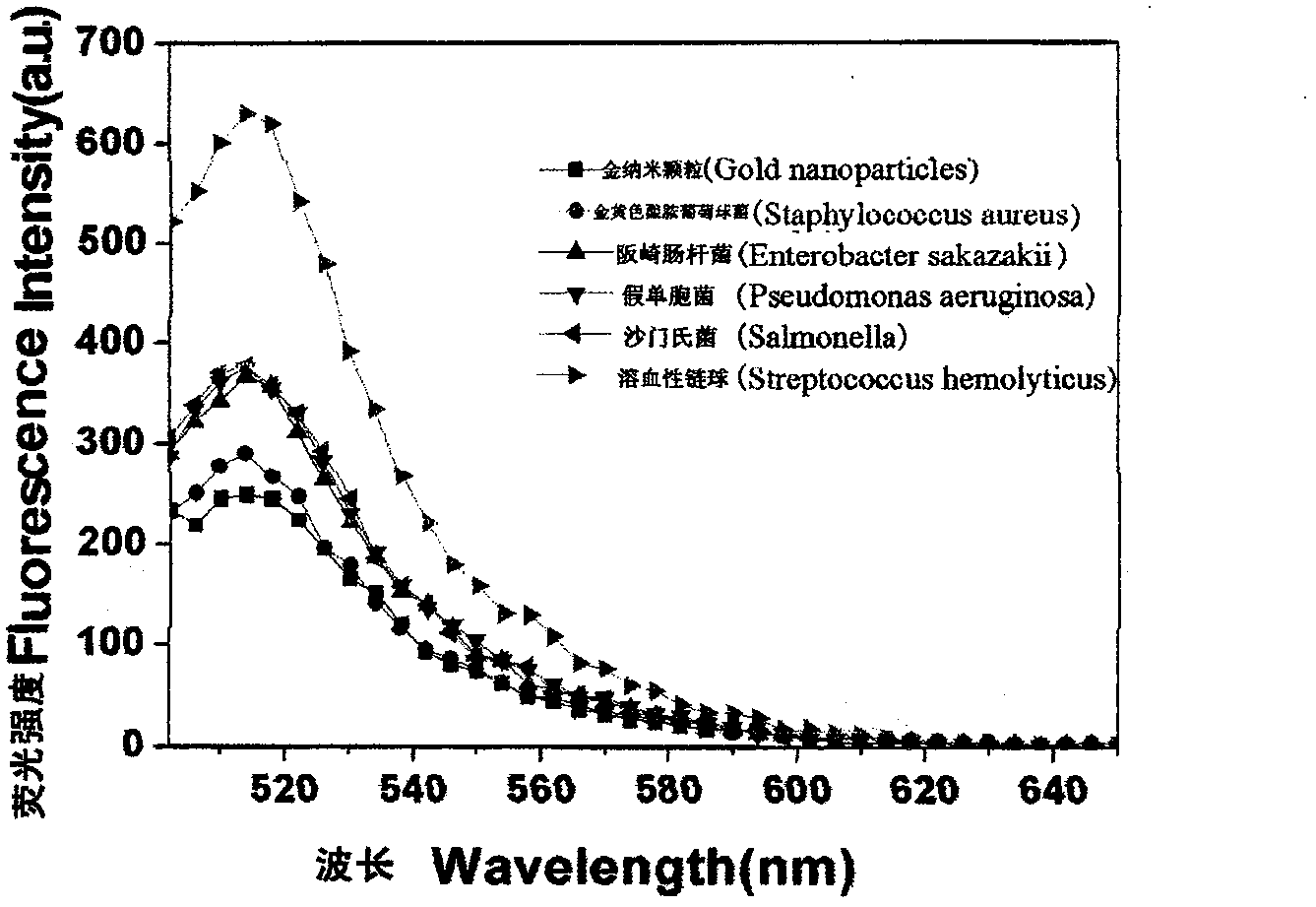
[0036] 1.3 Species: Hemolytic Streptococcus [CMCC (B) 32210], Staphylococcus aureus [ATCC6538], Salmonella [CMCC (B) 50115], Pseudomonas aeruginosa [CMCC (B) 10104], Bacillus sakazakii [ ATCC29004].
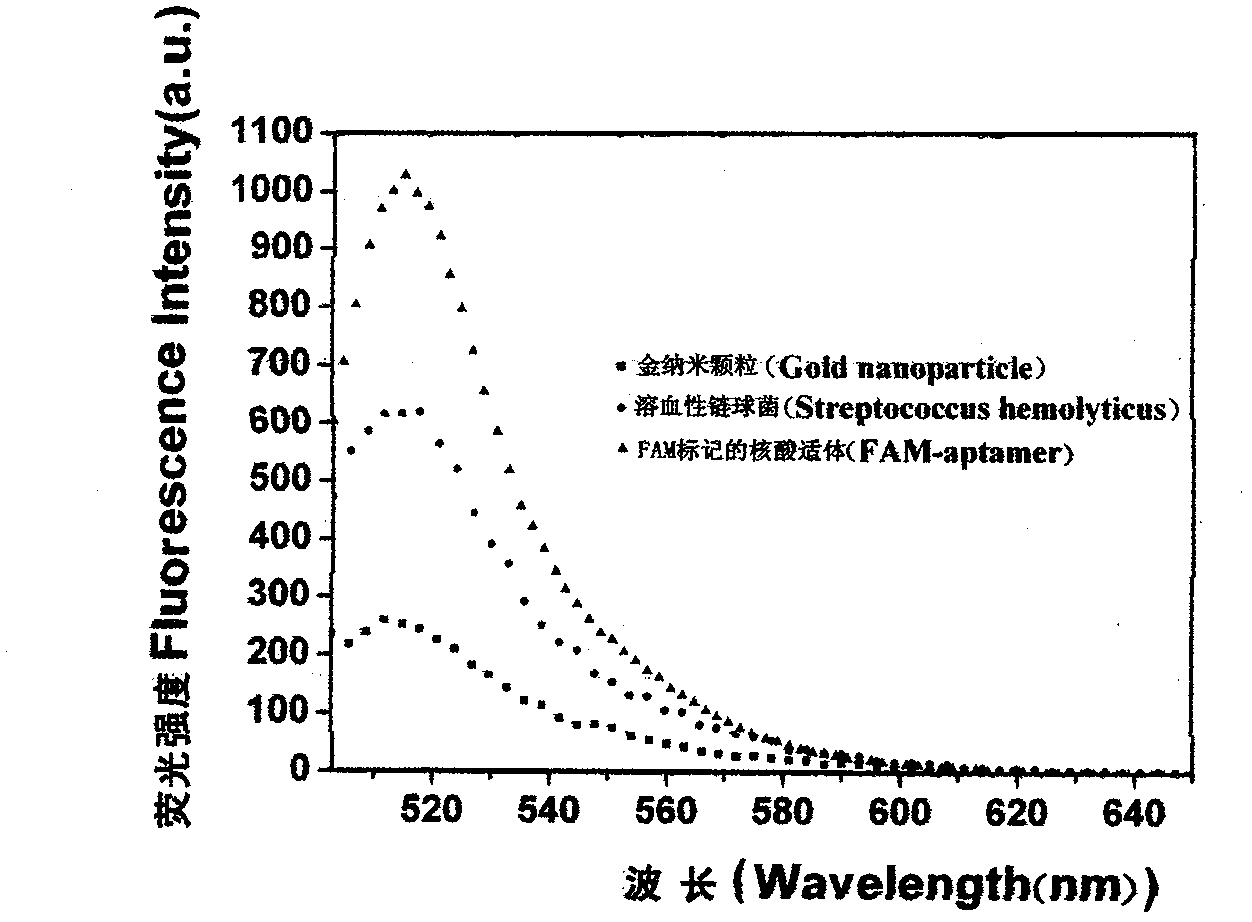
[0037] 2. A nucleic acid aptamer sequence marked with FAM that can specifically bind to hemolytic streptococcus was synthesized; at the same time, a complementary sequence of a DNA target sequence was synthesized.
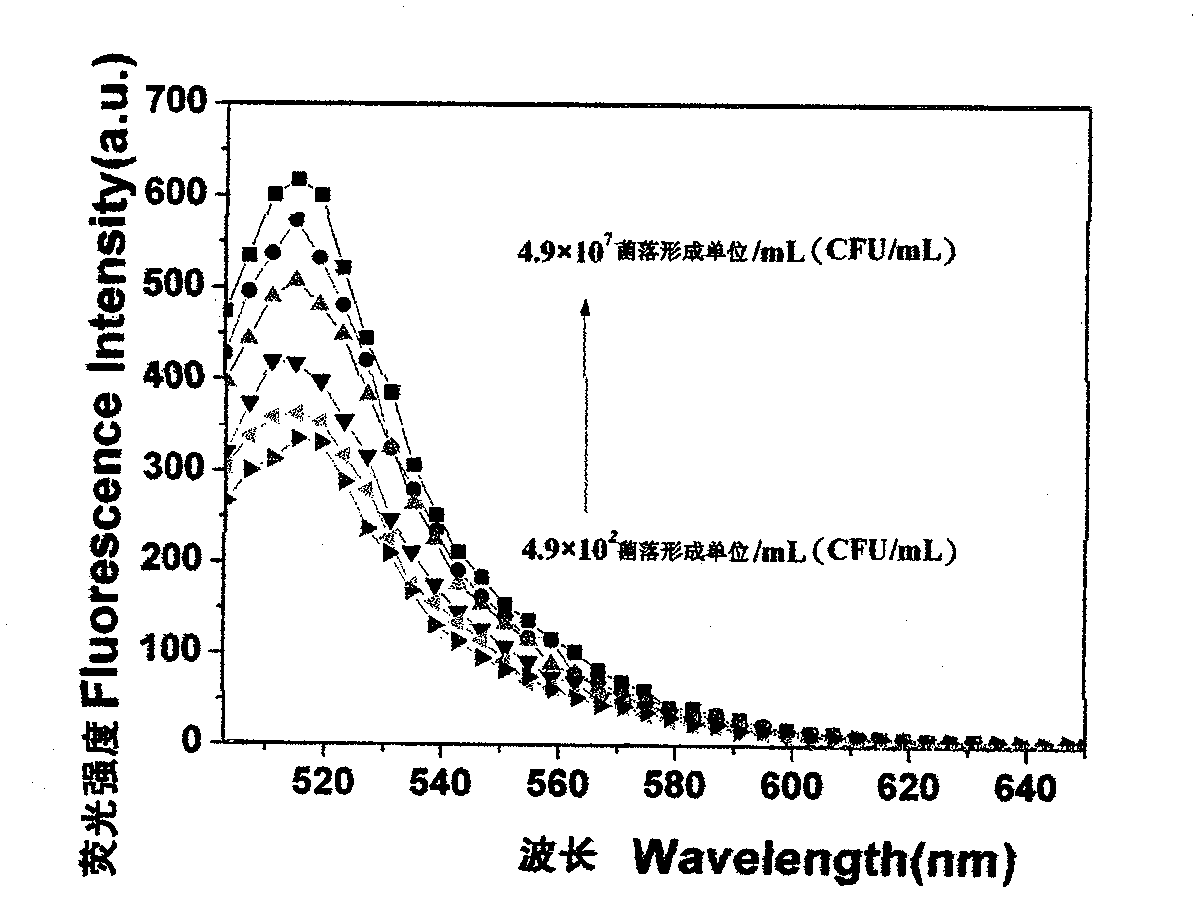
[0038] 3. Sample treatment: In the biological safety cabinet, the sample was diluted and enriched according to the requirements of GB4789.11-2003, and the sample enrichment solution was incubated in a 37°C incubator for...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com