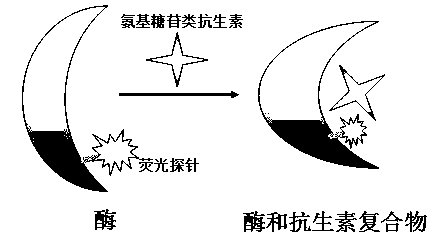
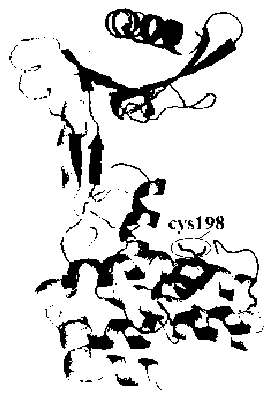Fluorescently-labeled aminoglycoside adenosine modification enzyme and method for detecting antibiotics with same
An aminoglycoside and glycoside adenosine technology, which is applied in the field of fluorescently labeled aminoglycoside adenosine modification enzymes and the detection of antibiotics, can solve the problems of interfering with the binding of antibiotics and ribosomes, and the loss of efficacy of antibiotics, and achieves enzymatic catalysis. Fast response, expensive reagents and consumables, and the effect of curbing application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The establishment of the standard curve of antibiotic concentration and fluorescent probe intensity: use a 494nm light source as the excitation light source, irradiate the sample cell of the quartz cuvette, then place the receiver in a vertical 90-degree direction, detect the emitted fluorescence, and record the data collection .
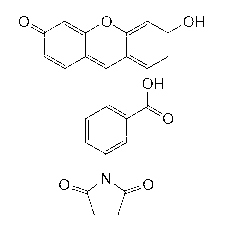
[0035] Sample preparation: Dissolve the known kanamycin A in neutral HEPES buffer (pH 7.5, 100mM KCl) at room temperature, and make successively different concentrations of standards (0.1μM, 0.5μM, 1μM, 5μM, 10μM, 20μM, 50μM, 100μM, 200μM, 500μM, 1mM). Blank experiment: First, on the free sulfhydryl Cys198 of the aminoglycoside adenosine modifying enzyme, fluorescein-5-maleimide (Fluorescein-5-maleimide) containing or modifying the sulfhydryl group is labeled, and its structure is as follows figure 2 shown. Then the labeled aminoglycoside adenosine modifying enzyme (ANT(4')), ATP, MgCl 2 Dissolve in neutral HEPES buffer (pH 7.5, 100mM KCl)...
Embodiment 2
[0046] Example 2: Detection of residual neomycin B in milk:
[0047] The problem of antibiotic residues in milk has become one of the hidden dangers of industry development and food safety. At the source of the dairy industry, antibiotics are used frequently, especially for the treatment of bovine mastitis, and are often used repeatedly in large doses. In dairy cow feed, antibiotic additives are also contained. Therefore, the problem of antibiotic residues in milk is very serious. This protocol can be applied to the rapid detection of aminoglycoside antibiotics in milk, taking neomycin B as an example. Take 5ml of milk (such as Yili) on the market, add a small amount of neomycin to make the concentration 10μM, and mix well. Then centrifuge on a high-speed centrifuge with a rotating speed of 21000 rpm for 2.5 hours. After centrifugation, the milk is separated, and large proteins are precipitated. Neomycin B has a relatively small molecular weight and remains in the supern...
Embodiment 3
[0048] Example 3: Detection of residual kanamycin A in milk:
[0049] This protocol can be applied to the rapid detection of aminoglycoside antibiotics in milk, taking kanamycin A as an example. Take 5ml of milk (such as Yili) on the market, add a small amount of kanamycin to make the concentration 6μM, and mix well. Then centrifuge on a high-speed centrifuge with a rotating speed of 21000 rpm for 2.5 hours. After centrifugation, the milk is separated, and large proteins are precipitated. Kanamycin A has a relatively small molecular weight and remains in the supernatant aqueous solution. Carefully transfer the aqueous supernatant to a new centrifuge tube for fluorescence testing. Then the reaction system was prepared, and the fluorescently labeled aminoglycoside adenosine modifying enzyme, ATP, and metal ions were configured in neutral HEPES buffer (pH 7.5, 100mM KCl) according to a certain concentration, wherein the modifying enzyme was 10 μM, and the ATP concentration wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com